Abstract
Viral infection outcomes are sex biased, with males generally more susceptible than females. Paradoxically, the numbers of antiviral natural killer (NK) cells are increased in males. We demonstrate that while numbers of NK cells are increased in male mice, they display decreased effector function compared to females in mice and humans. These differences were not solely dependent on gonadal hormones, because they persisted in gonadectomized mice. Kdm6a (which encodes the protein UTX), an epigenetic regulator that escapes X inactivation, was lower in male NK cells, while NK cell-intrinsic UTX deficiency in female mice increased NK cell numbers and reduced effector responses. Furthermore, mice with NK cell-intrinsic UTX deficiency showed increased lethality to mouse cytomegalovirus. Integrative multi-omics analysis revealed a critical role for UTX in regulating chromatin accessibility and gene expression critical for NK cell homeostasis and effector function. Collectively, these data implicate UTX as a critical molecular determinant of sex differences in NK cells.
Similar content being viewed by others
Main
Evolutionarily conserved sex differences exist in both innate and adaptive immune responses1,2. While males are less susceptible to autoimmunity, they also mount a less potent antiviral immune response than females3. For instance, males have a higher human cytomegalovirus (HCMV) burden after infection, suggesting increased susceptibility to viral threats4. This has also been recently illustrated during the coronavirus disease 2019 (COVID-19) pandemic, in which the strong male bias for severe disease has been postulated to reflect sex differences in immune responses5. Multiple studies in humans and mice have recently reported differences in immune cell distribution and/or function in males versus females6,7. However, the molecular basis for these differences, and the mechanisms by which these differences influence disease outcomes, remain poorly understood.
Sex differences in mammals are defined not only by divergent gonadal hormones, but also by sex chromosome dosage1. Expression of a subset of X-linked genes, for example, is higher in females (XX) than males (XY)8. While females undergo random X-chromosome inactivation (XCI) to maintain similar levels of X-linked protein expression between sexes, XCI is incomplete, with 3–7% of X-chromosome genes escaping inactivation in mice and 20–30% escaping inactivation in humans8,9. As such, differential levels of X-linked gene expression in females versus males have been linked to sex differences in a wide range of conditions including neural tube defects10 and autoimmune disease11,12.
As circulating type 1 innate lymphocytes, NK cells serve as an early line of defense against herpesvirus family members13. The importance of NK cells in antiviral immunity is illustrated in patients with defective NK cell numbers or functionality, who are highly susceptible to infection by herpesviruses such as HCMV and Epstein–Barr virus14. In mice, NK cells are required for the control of mouse cytomegalovirus (MCMV) and other viral infections15. Mice with either genetic deficiency in NK cell function or loss of NK cell numbers have a significant increase in viral titers and mortality following MCMV infection15,16,17,18. Thus, NK cells are critical in antiviral immunity in both mice and humans.
Given the potent antiviral function of NK cells, it was therefore unexpected that virus-susceptible males display higher numbers of NK cells6,7. Beyond NK cell numbers, other previously unappreciated sexually dimorphic NK cell feature(s) may instead account for sex differences during viral infection. We demonstrate that while male NK cells display enhanced cellular fitness in mice, they show decreased effector function in mice and humans. These sex biases in NK cell composition and function were not completely due to hormonal differences, because they persisted in gonadectomized mice. Through differential expression screening, we identified the X-linked epigenetic regulator and known XCI escapee UTX, which was expressed at significantly lower levels in both mouse and human male NK cells. UTX regulated both NK cell fitness and effector function in a dose-dependent manner, because UTX haploinsufficiency in female NK cells was sufficient to increase NK cell numbers while impairing cytokine production and cytotoxicity. Female UTX-deficient NK cells displayed enhanced persistence in vivo and resistance to apoptosis ex vivo, as well as increased susceptibility to MCMV infection. These effects were independent of UTX’s intrinsic demethylase activity, as NK cell numbers and interferon (IFN)-γ production were unaltered in mice expressing a ‘demethylase-dead’ UTX mutant. Integrative analysis using the assay for transposase-accessible chromatin using sequencing (ATAC-seq), bulk RNA sequencing (RNA-seq) and anti-UTX CUT&Tag (Cleavage Under Targets and Tagmentation Assay) of wild-type (WT) and UTX-deficient NK cells revealed a critical role for UTX in regulating expression of gene loci involved in NK cell fitness and effector responses. Our findings identify UTX as a major driver of sex differences in NK cell homeostasis and effector function through demethylase-independent modulation of gene expression.
Results
NK sexual dimorphism is independent of gonadal sex hormones
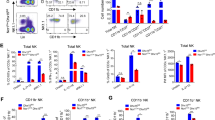
A recent investigation examining spleens of C57BL/6 mice reported increased numbers of NK cells in males versus females19. Consistent with these data, we observed that splenic NK cells (identified as CD3−TCRβ−NK1.1+; Extended Data Fig. 1a) are increased in frequency (Fig. 1a,b) and absolute numbers (Fig. 1c) in male C57BL/6 mice compared to females. These findings suggest that other sexually dimorphic features beyond NK cell numbers may account for increased male susceptibility to viral infections. In response to viral infection, NK cells are critical for early production of pro-inflammatory cytokines, particularly IFN-γ20,21,22. To test if sex differences exist in NK cell-intrinsic function, we compared effector cytokine production in NK cells isolated from female versus male mice ex vivo. Stimulation with the pro-inflammatory cytokines interleukin (IL)-12 and IL-15 resulted in lower IFN-γ production by male NK cells (Fig. 1d,e and Extended Data Fig. 1b). Similar results were observed in response to IL-12 and IL-18 (Extended Data Fig. 1c,d), suggesting a respective defect in male NK cell responsiveness to cytokine stimulation. Additionally, human NK cells (TCRβ−CD3−CD56+) isolated from peripheral blood mononuclear cells (PBMCs) activated with IL-12 and K562 leukemia cells resulted in lower percentage IFN-γ+ (Fig. 1f and Extended Data Fig. 1e) and IFN-γ mean fluorescence intensity (MFI; Fig. 1g) in male versus female NK cells. Thus, although NK cell numbers are increased, male NK cell effector cytokine production is consistently reduced in both mice and humans in response to pro-inflammatory cytokines induced during viral infection.
a–c, Representative dot plots (a), frequency (b) and absolute numbers (c) of splenic NK cells (CD3−TCRβ−NK1.1+) in female and male C57BL/6 mice (n = 15 per group). d,e, Percentage IFN-γ+ (d) and normalized IFN-γ MFI (e) of total splenic NK cells from female versus male mice cultured with no treatment (NT) or IL-15 (50 ng ml−1) and IL-12 (20 ng ml−1) for 4 h, normalized to MFI of female IL-15/IL-12 treatment (n = 8 per group). f,g, Percentage IFN-γ+ (f) and normalized IFN-γ MFI (g) of CD3−CD56+ female (n = 6) and male (n = 7) human NK cells cultured and stimulated with 10 ng ml−1 of IL-12 for 16 h in the presence of K562 cells, normalized to MFI of female IL-12 treatment. h,i, Frequency (h) and absolute numbers (i) of splenic NK cells in gonadectomized female and male mice (n = 18 per group). j,k, Percentage IFN-γ+ (j) and normalized IFN-γ MFI (k) of total splenic NK cells isolated from gonadectomized female and male mice and cultured with NT or IL-15 (50 ng ml−1) and IL-12 (20 ng ml−1) for 4 h (n = 12 per group). Data are representative of 2–4 independent experiments. Samples were compared using a two-tailed unpaired Student’s t-test and data points are presented as individual mice with the mean ± s.e.m. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Specific P values are as follows: b = 0.0002; c = 0.006; d = 0.0036; e = 0.0013; f = 0.04; g = 0.03; h < 0.0001; i = 0.0006; j = 0.0234; k = 0.0019.
Female or male sex is based on a composite of gonadal hormones (for example, estrogens or androgens) and sex chromosomes (for example, 46XX or 46XY)1. Previous studies demonstrated direct effects of gonadal hormones in regulation of IFN-γ production by NK cells23, but it remains possible that NK cell sex differences could also be attributed to cell-intrinsic factors. To identify sex hormone-mediated effects, we examined NK cell abundance and function in gonadectomized mice. Gonadectomy failed to eliminate sex differences in NK cell frequency (Fig. 1h and Extended Data Fig. 1f), absolute numbers (Fig. 1i) and IFN-γ protein production in response to cytokine stimulation (Fig. 1j,k and Extended Data Fig. 1g), indicating gonadal hormones are not solely responsible for sex differences in NK cells. While NK cell maturation subsets identified by CD11b and CD27 expression have differential capacities for survival and effector function24, splenic NK cells derived from either WT or gonadectomized female and male mice did not display significant differences in the frequencies of NK cell maturation subsets (Extended Data Fig. 1h–k). These results indicated that the observed sex biases in NK cell number and effector function are not due to differential maturation states. Thus, we hypothesized that sex chromosome dosage may contribute to differential NK cell abundance and function between sexes.
UTX escapes X inactivation and is expressed more in females
While 46XX females undergo XCI to control dosages of X-linked genes, a subset of genes escapes XCI (termed XCI escapees), often resulting in higher expression in females compared to males25,26. Thus, increased XCI escapee expression in females compared to males could potentially mediate sex differences in NK cells. While different genes escape X inactivation in humans and mice, five genes (XIST, DDX3X, KDM6A, EIF2S3, KDM5C) have previously been identified as XCI escapees in both27. XIST was excluded from further analysis because it is not expressed in male cells due to its known role in XCI in female cells1. All four remaining genes were significantly downregulated in male versus female NK cells, in both human (Fig. 2a) and mouse (Fig. 2b). Kdm6a (which encodes UTX) transcript levels displayed the most sexually dimorphic expression in both human and mouse NK cells (Fig. 2a,b). Male NK cells also expressed lower UTX protein levels compared to female NK cells in mice (Fig. 2c,d). These differences in Kdm6a transcript levels and UTX protein levels persisted in both gonadectomized mice (Fig. 2e–g) and Four Core Genotypes (FCG) mice (Extended Data Fig. 2a) in which sex chromosome complement (XX or XY) is uncoupled from gonadal sex organ (ovaries or testes)28. These data indicate expression levels of Kdm6a (UTX) are sex biased in NK cells and primarily dictated by X-chromosome dosage rather than gonadal hormones.
a, Normalized expression of XCI escapee genes using DICE database RNA-seq data on sorted NK cells from human females (n = 36) versus males (n = 54) normalized to female. b, Normalized expression of XCI escapee genes by quantitative PCR with reverse transcription (RT–qPCR) in splenic NK cells from female versus male mice (C57BL/6; 8 weeks old, n = 5 per group). Genes are ordered by increasing fold change between female and male from left to right. c,d, Representative histogram (c) and normalized MFI (d) of UTX protein expression in splenic NK cells from naive female versus male mice by flow cytometry, normalized to MFI of female mice (C57BL/6; 8 weeks old; n = 15 per group). e, Relative expression of Kdm6a (UTX) by RT–qPCR of isolated splenic NK cells normalized to female (n = 6 per group). f,g Representative histograms (f) and relative UTX MFI (g) of NK cells by flow cytometry from spleens of gonadectomized female and male mice (n = 6 per group) normalized to female. Samples were compared using unpaired two-tailed Student’s t-test and data points are presented as individual mice with the mean ± s.e.m. (*P < 0.05; **P < 0.01; ***P < 0.001). Specific P values are as follows: a < 0.001; b: Ddx3x = 0.03, Kdm5c = 0.0017, Eif2s3 = 0.0113, Kdm6a = 0.000087; d = 0.003; e = 0.008; g = 0.0029).
UTX suppresses NK cell fitness
To determine if UTX mediates the observed sex differences in NK cells, we generated a series of mice with dose-dependent loss of UTX. First, we generated female mice with a heterozygous deletion of UTX in NK cells (Kdm6afl/WT Ncr1Cre+, hereafter referred to as UTXHet; Supplementary Data Table 1) to mimic the single copy of UTX expressed in males. We confirmed similar NK cell UTX protein expression between female UTXHet and male WT (Kdm6afl/y Ncr1Cre-) mice (Extended Data Fig. 2b). Female UTXHet mice displayed similar splenic NK cell numbers compared to male WT (Fig. 3a,b), and both displayed increased numbers of NK cells compared to female WT. No significant differences in maturation by CD11b and CD27 expression were observed between NK cells from female WT, male WT and female UTXHet mice (Extended Data Fig. 2c). Thus, loss of one copy of UTX was sufficient to increase NK cell numbers in a maturation-independent manner.
a,b, Frequency (F and M WT: n = 12; F UTXHet: n = 16; a) and absolute numbers (F WT: n = 6; M WT: n = 8; F UTXHet: n = 9; b) of NK cells in spleen of female (F) WT, male (M) WT and F UTXHet mice. c,d, Frequency (WT: n = 12; UTXHet: n = 16; UTXNKD: n = 6; c) and absolute numbers (WT: n = 8; UTXHet: n = 12; UTXNKD: n = 6; d) of NK cells in spleen of F WT, UTXHet and UTXNKD mice. e, Representative contour plots of congenically distinct WT (CD451x2) and UTXNKD (CD45.2+) NK cells transferred into WT (CD45.1+) recipients at a 1:1 ratio before injection (left) and on day 7 after transfer (right). f, Frequency of WT and UTXNKD cells in the spleen of recipient mice before injection and day 7 after transfer (n = 6). g,h, Representative histograms (g) and percentage (h) of cleaved caspase 3+ NK cells of female WT, UTXHet and UTXNKD mice cultured with IL-15 (5 ng ml−1) and either dimethylsulfoxide (DMSO; F WT: n = 7; F UTXHet: n = 11; F UTXNKD: n = 6) or 2.5 μM Nutlin-3a (F WT: n = 3; F UTXHet: n = 7; F UTXNKD: n = 3) for 24 h. i–k, Normalized Bcl-2 MFI (i), Bim MFI (j) and Bcl-2:Bim MFI ratio (k) in splenic NK cells from female WT and UTXNKD mice (n = 5). l,m, Bcl-2:Bim MFI ratio in splenic NK cells from female WT and male WT mice (n = 6; l) and gonadectomized female and male mice (n = 11; m). Data are representative of 2–4 independent experiments. Samples were compared using ordinary one-way analysis of variance (ANOVA; a–d), two-way ANOVA with Tukey’s correction for multiple comparisons (h) or unpaired two-tailed Student’s t-test (f,i–m). Data points are individual mice with the mean ± s.e.m. (NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Specific P values are as follows: a: F WT versus M WT = 0.0201, M WT versus UTXHet = 0.989, F WT versus UTXHet = 0.0327, WT versus UTXNKD = 0.001; b: F WT versus M WT = 0.0320, M WT versus UTXHet < 0.99, F WT versus UTXHet = 0.0029, WT versus UTXNKD = 0.001; c: WT versus UTXHet = 0.0375, UTXHet versus UTXNKD and WT versus UTXNKD < 0.0001; d: WT versus UTXHet = 0.0191, UTXHet versus UTXNKD = 0.0278, WT versus UTXNKD = 0.001; f: Pre-injection = 0.3304; day 7 = 0.001918; h: DMSO < 0.0001, Nutlin-3a–F WT versus F UTXHet = 0.0048, rest < 0.0001; i < 0.0001; j = 0.0004; k = 0.0025; l = 0.0115; m = 0.0227.
We next produced mice with a homozygous deletion of UTX (Kdm6afl/fl Ncr1Cre+, hereafter referred to as UTXNKD; Supplementary Data Table 1), resulting in loss of both copies of UTX in NK cells. UTX protein expression was significantly lower in female UTXNKD NK cells compared to female WT NK cells by flow cytometry (Extended Data Fig. 2b), as well as lower compared to NK cells with a single UTX copy (that is, male WT and female UTXHet). The absence of UTX protein at the predicted size (180 kD) in female UTXNKD compared to WT NK cells was confirmed by western blot (Extended Data Fig. 2d). NK cell frequencies and absolute numbers increased with decreasing UTX copy number (Fig. 3c,d and Extended Data Fig. 3a). These data implicate UTX in regulating NK cell frequency and absolute numbers in a dose-dependent manner.
To define the mechanisms underlying the increased NK cell numbers in UTXNKD mice (CD45.2+), a 1:1 ratio of mixed bone marrow chimeric (mBMC) mice with WT (CD45.1+) were produced. Six weeks after reconstitution, we observed a marked competitive advantage of female UTXNKD (CD45.2+) NK cells compared to WT in female recipients (CD451x2) following bone marrow transplantation (Extended Data Fig. 3b,c). In contrast to NK cells, T cells from the same donor (UTXNKD,CD45.2+), which are UTX sufficient due to NK-specific deletion of UTX, displayed the original injection ratio (1:1; Extended Data Fig. 3b,c). These data suggest UTX repressed NK cell numbers in a cell-intrinsic manner during development. To test whether this phenotype was driven by differences in proliferation, we analyzed the cell division marker Ki67 in splenic NK cells in WT:UTXNKD mBMC mice, injected at a 4:1 ratio to normalize cell number between genotypes. Paradoxically, UTXNKD NK cells displayed lower frequencies of Ki67+ cells and showed less CFSE dilution in response to IL-15 (Extended Data Fig. 3d,e). These results suggest that the higher NK cell numbers observed in UTXNKD mice were not due to increased proliferation.
Given these results, we hypothesized that the elevated frequency of NK cells in UTXNKD mice (Fig. 3c,d) could be due to enhanced cellular fitness of NK cells in the absence of UTX expression. To test this possibility, congenically distinct WT (CD451x2) and UTXNKD (CD45.2+) splenic NK cells were labeled with Cell Trace Violet (CTV) and transferred into WT (CD45.1+) recipients at a 1:1 ratio (Extended Data Fig. 3f). On day 7 after transfer, the transferred population was skewed toward UTXNKD NK cells in recipient spleens (Fig. 3e,f), demonstrating cell-intrinsic UTX suppression of mature NK cell homeostasis. This difference was not due to altered proliferation, because CTV dilution by both transferred populations was minimal on day 7 after transfer (Extended Data Fig. 3g). To test whether UTX repressed NK cell homeostasis through regulation of apoptosis, we compared cleaved caspase 3 expression in sorted NK cells incubated with either IL-15 alone or with IL-15 and an apoptosis inducer, Nutlin-3a29. Lower UTX expression correlated with decreased cleaved caspase 3+ NK cells in the presence of low-dose IL-15 and Nutlin-3a treatment (Fig. 3g,h). Moreover, male NK cells also displayed a modest but significant decrease in the frequency of cleaved caspase 3+ NK cells in response to Nutlin-3a compared to female NK cells, which also persisted in gonadectomized mice (Extended Data Fig. 3h–k). Moreover, regulation of NK cell apoptosis and survival relies on the relative expression levels of Bcl-2 (anti-apoptotic factor)30, which can be antagonized by Bim (pro-apoptotic factor)31. UTXNKD NK cells showed increased intracellular protein expression of Bcl-2 and a modest increase in Bim (Fig. 3i,j and Extended Data Fig. 3l) compared to WT NK cells. This resulted in a significantly increased Bcl-2:Bim ratio in UTXNKD NK cells (Fig. 3k). Male NK cells also displayed a significant increase in Bcl-2:Bim ratio (Fig. 3l), which persisted following gonadectomy (Fig. 3m). Together, these data demonstrate that altered UTX levels may underlie sex differences in NK cell fitness through regulation of Bcl-2 expression.
UTX enhances NK cell effector function
Because male NK cells exhibited decreased IFN-γ production (Fig. 1d,e) independent of gonadal hormones (Fig. 1j,k) we next sought to determine if this phenotype was regulated by UTX levels. After cytokine stimulation, frequencies and absolute numbers of IFN-γ-producing cells (Fig. 4a and Extended Data Fig. 4a,b) as well as IFN-γ MFI (Fig. 4a) were similar between male WT and female UTXHet NK cells. IFN-γ production by female UTXHet NK cells was intermediate between female WT and female UTXNKD NK cells (Fig. 4a and Extended Data Fig. 4a,b). This trend was also observed by comparing NK cell IFN-γ accumulation by ELISA (Fig. 4b). Moreover, this phenomenon was not specific to IFN-γ, because granulocyte-macrophage colony-stimulating factor (GM-CSF) production, a pro-inflammatory NK effector molecule32, was also reduced with decreasing UTX copy number in female NK cells (Fig. 4c).
a, Percentage IFN-γ+ and normalized IFN-γ MFI of NK cells from female WT (n = 8), male WT (n = 8), female UTXHet (n = 12) and female UTXNKD (n = 6) mice with no treatment (NT) or IL-15 (50 ng ml−1) and IL-12 (20 ng ml−1) for 4 h, normalized to MFI of female IL-15/IL-12 treatment. b,c, IFN-γ (b) and GM-CSF (c) concentrations measured by ELISA in supernatants from female WT (n = 4), UTXHet (n = 5) and UTXNKD (n = 3) NK cells with NT or IL-12 (20 ng ml−1) and IL-18 (10 ng ml−1) for 4 h. d, Specific lysis of MHCI-deficient MC38 (target) cells by female WT (n = 5), male WT (n = 8), female UTXHet (n = 12) or female UTXNKD (n = 6) NK (effector) cells for 16 h at a 4:1 effector:target ratio, normalized to lysis by female WT. e, Kaplan–Meier survival curves of WT and UTXNKD mice infected with MCMV (n = 8). Mantel–Cox test (P = 0.0093). f, Percentage IFN-γ+ (n = 14), normalized IFN-γ MFI (n = 14) and granzyme B (GzmB) MFI (n = 8) relative to WT in splenic NK cells on D1.5 after MCMV infection of 4:1 WT:UTXNKD mBMC mice. g, Schematic of 1:1 WT (CD45.1+) and iUTX−/− (CD45.2+) bone marrow (BM) transferred into busulfan-depleted hosts and treated with 1 mg tamoxifen for 3 d before MCMV infection. h, Percentage IFN-γ+ and normalized IFN-γ MFI of NK cells from 1:1 WT:iUTX−/− mBMC mice on D1.5 after MCMV infection, normalized to WT (n = 6). i, Two-tailed Pearson correlation of IFN-γ versus UTX MFI of NK cells (n = 12; r2 = 0.775; P < 0.0001). Data are representative of 2–3 independent experiments. Two-way ANOVA (a–c), one-way ANOVA with Tukey’s correction for multiple comparisons (d) or paired two-tailed Student’s t-test (f,h) were used. Data points are individual mice with the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Specific P values: a: F WT versus M WT = 0.0027, F WT versus F UTXHet and F WT versus F UTXNKD < 0.0001, M WT versus F UTXHet = 0.269, F UTXHet versus F UTXNKD = 0.0007; b: WT versus UTXHet = 0.0037, UTXHet versus UTXNKD = 0.0011, WT versus UTXNKD < 0.0001; c: WT versus UTXHet = 0.0026, UTXHet versus UTXNKD = 0.0349, WT versus UTXNKD < 0.0001; d: F WT versus M WT = 0.0005, F WT versus F UTXHet and F WT versus F UTXNKD < 0.0001, M WT versus F UTXHet = 0.1616, F UTXHet versus F UTXNKD = 0.0439; f: %IFN-γ+ < 0.0001, IFN-γ MFI = 0.0024, GzmB = 0.0032; g: %IFN-γ+ < 0.0001, IFN-γ MFI = 0.0018. PI, post infection.
In addition to cytokine production, NK cell cytolytic activity is crucial for antiviral33 and antitumor defenses34. To assess sex differences in NK cell cytotoxicity, we performed killing assays with major histocompatibility complex (MHC) class I-deficient MC38 cells as targets. At a 4:1 effector:target ratio, male WT NK cells displayed significantly lower lysis of target cells compared to female WT (Fig. 4d), which was sustained in gonadectomized mice (Extended Data Fig. 4c). Impaired target cell killing by male NK cells was not due to differences in degranulation, because CD107a levels were similar between female and male NK cells (Extended Data Fig. 4d,e). However, males produced significantly lower levels of cytotoxic molecules perforin and granzyme B in response to IL-15 and anti-NK1.1 activating receptor ligation (Extended Data Fig. 4d,e). Notably, UTXHet and male WT NK cells showed similar killing capacity (Fig. 4d), which was intermediate between female WT and UTXNKD NK cells (Fig. 4d). Together, these data suggest that UTX enhances NK cell cytotoxicity in a dose-dependent manner.
Considering the observed effects of UTX loss on NK cell effector function, we examined whether UTXNKD mice were more vulnerable to viral infection. Rapid IFN-γ and GM-CSF production is critical for NK cell-mediated antiviral control20. Strikingly, UTXNKD mice rapidly succumbed to infection (n = 3/8 survived) upon challenge with a sublethal dose of MCMV (Fig. 4e). Moreover, UTX-deficient splenic NK cells displayed a marked defect in IFN-γ production and granzyme B production in total NK cells on day 1.5 after infection (Fig. 4f and Extended Data Fig. 4f,g). Additionally, a similar defect in IFN-γ production by UTXNKD was observed in all maturation subsets (Extended Data Fig. 4h), implicating UTX in control of IFN-γ production in a maturation-independent manner. To confirm whether dosage of UTX expression in mature NK cells associates with production of IFN-γ during viral infection in vivo, we generated transgenic mice to achieve a tamoxifen-inducible UTX deletion (Kdm6afl/fl Rosa26ERT2CRE+, hereafter referred to as iUTX−/−; Supplementary Data Table 1). mBMC mice were produced with a 1:1 mix of WT (CD45.1+) and iUTX−/− (CD45.2+) to limit UTX deletion to the hematopoietic compartment. WT:iUTX−/− mBMC mice were treated with tamoxifen immediately before infection with MCMV to ablate UTX expression (Fig. 4g). iUTX−/− (CD45.2+) NK cells produced less IFN-γ compared to their WT counterparts (Fig. 4h and Extended Data Fig. 4i). Tamoxifen administration in WT:iUTX−/− mBMC mice resulted in differential degrees of UTX protein loss and displayed a significant positive correlation between intracellular UTX levels and IFN-γ production on day 1.5 after infection (Fig. 4i). These results demonstrate that cell-intrinsic UTX levels in mature NK cells regulate effector molecule production and subsequent protection against MCMV infection.
UTX regulates NK cells in a demethylase-independent manner
As a histone demethylase, UTX may control NK cell homeostasis and effector gene expression programs by catalyzing the removal of a methyl group from trimethylated histone H3 Lys27 (H3K27me3; a repressive histone mark) to poise chromatin for active gene expression35. However, UTX also possesses demethylase-independent activities by interacting with epigenetic regulators and chromatin modifiers to coordinate gene expression36,37. To explore the role of UTX demethylase activity in modulating NK cell homeostasis and effector function, we leveraged mice that express a catalytically inactive UTX (UTX ‘demethylase-dead’ or UTXDMD mice; Supplementary Data Table 1) harboring p.His1146Ala and p.Glu1148Ala point mutations in the catalytic domain38. Interestingly, female UTXDMD and WT mice exhibited similar frequencies and absolute numbers of splenic NK cells (Fig. 5a–c), while UTXNKD mice showed increased splenic NK cell numbers compared to both. These findings suggest that UTX’s repression of NK cell numbers was demethylase independent. Moreover, no differences were observed between WT and UTXDMD NK cells in the ability to produce IFN-γ in response to cytokine stimulation (Fig. 5d–f). These results demonstrate that UTX’s function in restraining NK cell numbers and promoting IFN-γ production is demethylase independent.
a–c, Representative density plots (a), frequency (b) and absolute number (c) of NK cells in the spleens of female WT, UTXDMD and UTXNKD mice (n = 5 per group). d–f, Representative contour plots (d), percentage IFN-γ+ (e) and normalized IFN-γ MFI (f) of female WT, UTXDMD and UTXNKD NK cells (n = 5 per group) normalized to WT. Data are representative of 2–3 independent experiments. Samples were compared using one-way ANOVA with Tukey’s correction for multiple comparisons. Data points are presented as individual mice with the mean ± s.e.m. *P < 0.05; **P < 0.01; ****P < 0.0001. Specific P values are as follows: b: WT versus UTXDMD = 0.7353, WT versus UTXNKD and UTXDMD versus UTXNKD < 0.0001; c: WT versus UTXDMD = 0.4888, WT versus UTXNKD and UTXDMD versus UTXNKD < 0.0001; e: WT versus UTXDMD = 0.855, WT versus UTXNKD = 0.0030, UTXDMD versus UTXNKD = 0.0380; f: WT versus UTXDMD = 0.1382, WT versus UTXNKD = 0.0079, UTXDMD versus UTXNKD = 0.0166.
In addition to a single copy of UTX, males express the catalytically inactive Kdm6c (which encodes the protein UTY), the Y-chromosome-linked homolog of UTX. We confirmed that UTY is only expressed in male NK cells from humans (Extended Data Fig. 5a) and mice (Extended Data Fig. 5b) and was not altered by gonadectomy in mice (Extended Data Fig. 5b). As discussed above, no differences were seen in NK cell numbers or effector function between male WT (one copy of UTX and UTY) and female UTXHet (one copy of UTX) mice (Figs. 3a,b and 4a,d), suggesting a limited role for UTY in regulating NK cell homeostasis and effector function. Additionally, male UTXNKD (Kdm6afl/y Ncr1cre+) mice showed increased NK cell frequency and absolute numbers (Extended Data Fig. 5c,d) and produced less IFN-γ (Extended Data Fig. 5e–h), compared to male WT controls, which mirrors changes seen in females with NK cell UTX deficiency. (Figs. 3a,b and 4a,b). Thus, UTX loss in NK cells has similar effects in females and males.
UTX controls NK cell transcriptome by remodeling chromatin
Recent studies have identified NK cell regulatory circuitry (regulomes) that prime innate lymphoid cells for swift effector responses even before NK cell activation39. As an epigenetic modifier, UTX can alter transcription by organizing chromatin at regulatory elements of target gene loci40. To investigate the UTX-mediated modifications on chromatin accessibility and gene expression in NK cells, we performed ATAC-seq in tandem with bulk RNA-seq on sort-purified WT (CD45.1+) and UTXNKD (CD45.2+) NK cells from 4:1 WT:UTXNKD mBMC mice (Extended Data Fig. 6a). Using mBMC mice allowed for an internally controlled experiment and minimized environmental confounding factors. Principal-component analysis (PCA) of both ATAC-seq and RNA-seq data revealed sample clustering by genotype (Extended Data Fig. 6b). ATAC-seq revealed 3,569 peaks decreased and 2,113 peaks increased in accessibility in UTXNKD compared to WT NK cells (log2 fold change > ±0.5, adjusted P value < 0.05, false discovery rate (FDR) < 0.05; Supplementary Data Table 2). Moreover, RNA-seq identified 701 decreased and 554 increased genes in UTXNKD versus WT (log2 fold change > ±0.5, adjusted P value < 0.05, FDR < 0.05; Extended Data Fig. 6c and Supplementary Data Table 3). These findings suggest profound changes in both the chromatin landscape and transcriptome of NK cells in the absence of UTX.
Integrative analysis of ATAC-seq and RNA-seq identified 395 genes that are both differentially accessible and expressed with a significant positive correlation (Spearman correlation: R = 0.62, P < 2.2 × 10−16) between the mean log2 fold change of ATAC-seq peaks and log2 fold change of RNA-seq expression (Extended Data Fig. 6d). Fuzzy c-means clustering of both the ATAC-seq and RNA-seq datasets identified six major clusters that were significantly decreased (clusters 1, 2, 3 and 6) or increased (clusters 4 and 5) in accessibility (Fig. 6a) and expression (Fig. 6b) in UTXNKD NK cells. For functional enrichment analysis, g:Profiler was used to analyze clusters of differentially expressed genes identified by RNA-seq (Fig. 6c). Major pathways such as immune system process, cytokine production, IFN-γ production, lymphocyte activation and immune effector process were associated with decreased expression in UTXNKD (clusters 1, 2, 3 and 6; Fig. 6c). Meanwhile, pathways such as developmental process, biosynthetic process and metabolic process were significantly associated with increased expression in UTXNKD (clusters 4 and 5; Fig. 6c). Of note, analysis of cell death pathway genes revealed multiple genes to be differentially expressed (Extended Data Fig. 6e). Notably, expression of the anti-apoptotic gene Bcl2 was increased, while the pro-apoptotic gene Casp3 was decreased (Extended Data Fig. 6e). Collectively, these findings implicate loss of UTX results in modifications of chromatin accessibility and expression of genes associated with NK cell homeostasis and effector function.
a–c, 4:1 WT:UTXNKD mBMCs were generated by transferring WT (CD45.1+) and UTXNKD (CD45.2+) bone marrow into lymphodepleted host mice (CD451x2) and allowed to reconstitute for 6 weeks. Splenic NK cells were sorted for ATAC-seq and RNA-seq library preparation (n = 3 per group). Line graphs (left) and heat map (right) of fuzzy c-means clustered differentially accessible peaks identified by ATAC-seq (a) and differentially expressed genes identified by RNA-seq (b) of splenic NK cells from WT:UTXNKD mBMC mice (adjusted P value < 0.05 and membership score > 0.5). Line graphs show mean (black line) and standard deviation (red ribbon) of mean-centered normalized log2 values of significant (FDR and adjusted P value < 0.05). c, Pathway analysis of significant fuzzy c-means clustered RNA-seq genes using g:Profiler with point size indicating −log10(P value; calculated by g:GOSt using Fisher’s one-tailed test). d–g, Anti-UTX CUT&Tag was performed in WT and UTXNKD NK cells and identified 5,746 unique UTX-bound peaks (n = 3 per group). d, Venn diagram outlining overlapping differentially accessible (DA) genes identified by ATAC-seq and differentially expressed (DE) genes identified by RNA-seq. e, Location of UTX-bound peaks. f,g, Representative gene tracks from UCSC Integrated Genome Browser of anti-UTX CUT&Tag (‘anti-UTX’), ATAC-seq and RNA-seq of Bcl2 and Thy1 (f) and Ifng and Csf2 (g); y axis depicts counts per million (CPM). h, Schematic of how differential UTX expression levels underlie sexual dimorphism in NK cell composition and function (left). Diagram of how UTX may be regulating gene programs involved in NK cell numbers and effector function during homeostasis and viral infection (right). Created with BioRender.com.
Considering the genome-wide differences in accessibility and gene expression we observed by ATAC-seq and RNA-seq, we also explored direct UTX-mediated effects. We performed anti-UTX CUT&Tag followed by sequencing, allowing detection of DNA regions bound to UTX using an antibody-based immunoprecipitation method41. Anti-UTX CUT&Tag on sort-purified WT and UTXNKD NK cells revealed 5,746 UTX-bound peaks (FDR < 0.01, adjusted P value < 0.05; Fig. 6d and Supplementary Data Table 4). PCA of both ATAC-seq and RNA-seq data revealed sample clustering by genotype (Extended Data Fig. 6f). We identified 191 genes that were UTX bound, differentially accessible by ATAC-seq and differentially expressed by RNA-seq (Fig. 6d). Within these 191 genes, a majority of UTX-bound peaks were located in promoter (20.58%), intronic (46.94%) and intergenic (27.4%) regions (Fig. 6e). Noteworthy genes involved in NK cell homeostasis (Bcl2 and Thy1; Fig. 6f)30,42 and effector function (Ifng and Csf2) were UTX bound, differentially accessible and differentially expressed (Fig. 6g). Moreover, of the 191 UTX-bound genes, 140 genes were decreased in expression, while the remaining 51 genes were increased (Supplementary Data Table 3) corroborating a prior report in T cells that UTX functions in both activating and repressing gene transcription43. Enrichr pathway analysis44 on these 191 UTX-bound genes revealed decreased inflammatory response, IFN-γ signaling and NK cell cytotoxicity pathways in UTXNKD NK cells (Extended Data Fig. 6g). Conversely, increased cellular catabolic process and apoptosis signaling pathways, which include both pro-apoptotic and anti-apoptotic genes (for example, Bcl2, Bbc3 and Gadd45g), were seen in UTXNKD NK cells. Among 865 UTX-bound peaks with UTX-dependent expression and chromatin accessibility differences (191 unique genes), linear regression analysis showed a significant positive correlation (Pearson’s R = 0.5165, P < 0.0001) between chromatin accessibility and gene expression (Extended Data Fig. 6h,i). UTX occupied regions within the Bcl2, Thy1, Ifng and Csf2 gene loci corresponded with regions in which differences in accessibility and gene expression were also noted (Fig. 6f,g). These data suggest UTX regulates chromatin accessibility and gene transcription pathways important in regulating NK cell homeostasis and function.
UTX is known to interact with transcription factors (TFs) to orchestrate target gene transcription40. To identify putative TF motifs with differential accessibility due to loss of UTX, we performed HOMER (Hypergeometric Optimization of Motif Enrichment)45 TF motif analysis on differentially accessible peaks identified by ATAC-seq (Extended Data Fig. 6j). TFs associated with NK cell effector function (for example, Runt (Runx1 and Runx2)46 and T-box (Eomes, T-bet, Tbr1 and Tbx6)47 family members) were more significant and had a higher percentage of target motifs associated with decreased accessibility in UTXNKD (clusters 1, 2, 3 and 6; Extended Data Fig. 6j). Conversely, TFs associated with proliferation, differentiation and metabolism in the zinc finger and ETS family TFs48 were more significantly associated with increased accessibility (clusters 4 and 5; Extended Data Fig. 6j). Furthermore, TF motif analysis of UTX-bound peaks by UTX CUT&Tag corroborates these results by revealing TFs critical in both NK cell effector processes (T-bet, Eomes, Runx1 and Tbx5) and developmental programs (ETS1 and AP-1; Extended Data Fig. 6k). These data suggest both differential accessibility and direct UTX binding of important TF binding motifs implicated in regulating NK cell fitness and effector processes. These analyses suggest that UTX modulates the chromatin landscape to control expression of genes important in NK cell homeostasis (Bcl2 and Thy1) and effector function (Ifng and Csf2). Ultimately, these findings suggest a model in which differential UTX expression levels may underlie sexual dimorphism in NK cells as a central regulator of NK cell fitness and effector function (Fig. 6h).
Discussion
Sex is a critical biological variable in determining outcomes to viral infections3. This was recently illustrated with COVID-19, in which male sex was identified as a major risk factor for severe disease5. Moreover, recent studies have linked NK cell dysfunction to severe COVID-19 disease49. Given the importance of NK cells in antiviral immunity, understanding the root causes of sex differences in NK cell biology will have far-reaching implications in optimizing endogenous effector responses. In this study, we demonstrate that lower UTX expression in male NK cells contributes to their increased numbers and decreased effector functionality. NK cell UTX is required for controlling NK cell fitness, modulating accessibility of TF binding motifs, increasing chromatin accessibility at effector gene loci and poising NK cells for rapid response to viral infection.
In addition to NK cells, sexual dimorphism has been reported in B cells, monocytes, neutrophils, CD4+ T cells and CD8+ T cells25. While sex differences in immune cells have previously been reported to be mediated by gonadal sex hormones50,51,52, it remains possible that a subset of these disparities may also be attributed to differential UTX expression. In support of this possibility, UTX deficiency has been associated with decreased T cell and invariant NK T cell numbers53,54,55; and UTX transcripts are lower in male versus female cells for multiple immune cell types (CD4+ T cells, CD8+ T cells, monocytes, B cells) queried in the DICE database56. Further phenotypic studies are needed to determine UTX’s role in modulating sex differences in other immune cell types.
NK cell-mediated effector functions include cytokine production and cytotoxic molecule expression. Our multi-omic analyses suggest that UTX poises the chromatin landscape of NK cells to quickly respond to viral challenge by increasing accessibility and transcription of effector loci. These studies revealed 191 genes (including Ifng and Csf2)20,32 that were simultaneously bound by UTX, differentially accessible and expressed. Decreased IFN-γ and GM-CSF cytokine production and impaired cytolytic capacity of UTX-deficient NK cells support UTX’s role in promoting effector functionality during inflammation. However, there may be additional indirect consequences of UTX-mediated gene regulation that play important roles in NK cell effector function, as evidenced by the additional genes that are differentially expressed but not bound by UTX.
As a H3K27me3 demethylase, UTX poises chromatin for active gene expression57. In addition to its catalytic activity, UTX functions in multiprotein complexes with other epigenetic regulators (for example, SWI/SNF, MLL4/5 and p300) to mediate chromatin remodeling in a demethylase-independent manner36,57. We report demethylase-independent functions of UTX in regulating homeostasis and effector programs in NK cells. This is in contrast to UTX’s role in invariant NK T cells, in which its demethylase activity is required53. Thus, the molecular mechanisms by which UTX functions may be lineage specific. In support of this hypothesis, UTX has been reported to interact with lineage-specific TFs in T cells to target effector loci40. Our HOMER motif analysis revealed potential UTX interactions with Runx1, Runx2, Eomes and other TFs important for NK cell effector function during viral infection46,47. Moreover, these analyses also point to UTX interactions with KLF1, KLF5, Sp2 and other TFs associated with NK cell proliferation, differentiation and metabolism48. Furthermore, our results support previously published studies in which UTX in other cell types has been reported to coordinate responses with T-bet, Eomes and Tbx5 (ref. 40), ETS1 (ref. 48) and AP-1 (ref.58). Finally, Runx1 has been shown to interact with UTX-regulated BRG1 and SWI/SNF complexes46. However, due to the correlative nature of HOMER analysis, further studies with co-immunoprecipitation and mass spectrometry are needed to experimentally verify these interactions in situ. Furthermore, as a constitutive XCI escapee, UTX may have cell-type-specific mechanisms through two possibilities: (i) pool of cofactors present and (ii) availability of its epigenetic binding partners. UTX relies on byproducts of metabolic pathways for cofactors (for example, Fe(II), α-ketoglutarate and oxygen) crucial for its enzymatic activity59. Thus, dependent on the metabolic state, there are distinct pools of cofactors accessible for UTX’s demethylase function, allowing differential levels of catalytic activity based on the cell type. Additionally, UTX’s functionality may also be contingent on the activity and expression level of its binding partners (for example, MLL3/4, SWI/SNF and p300) which are autosomally encoded.
Weighing factors that define patient subsets with different immune responses will allow us to move past a ‘one-size-fits-all’ therapeutic approach to a precision medicine paradigm. UTX deficiency has been associated with Kabuki syndrome and Turner syndrome60, two human conditions associated with immune dysregulation and increased infections. Our findings suggest the possibility that UTX deficiency in human NK cells may contribute to decreased viral immunosurveillance observed in these patients, although future work will be needed to support this hypothesis. Moreover, understanding sex differences in NK cell function is required to incorporate sex as a biological factor in treatment decisions. In males with severe viral illness, for instance, enhancing NK cell UTX activity may provide therapeutic benefit. We expect that these insights will be important not only in the setting of viral infections, but also in other infections and cancer, where NK cells also play an important role. These findings may also have important implications for adoptive cellular therapies, in which NK cells are the subject of intense interest61.
Methods
Mice
Mice were bred at UCLA in accordance with the guidelines of the Institutional Animal Care and Use Committee. The following mouse strains were used in this study: C57BL/6 (CD45.2+; Jackson Labs, 000664), B6.SJL (CD45.1+; Jackson Labs, 002114), Rosa26ERT2Cre (Jackson Labs, 008463), Ncr1Cre (ref. 62), Kdm6afl/fl (ref. 63), UTXH1146A+E1148A (UTXDMD)38 and FCG mice28 (B6.Cg-Tg(Sry)2Ei Srydl1Rlb/ArnoJ; Jackson, 10905). For experiments with gonadectomy, the procedure was performed by Jackson Laboratories Surgical Services. To generate UTXNKD and UTXHet mice de novo for this study, Ncr1Cre/WT mice were crossed to Kdm6afl/fl mice to generate either Ncr1Cre/WTKdm6afl/fl (UTXNKD) or Ncr1Cre/WTKdm6afl/WT (UTXHet) offspring. To generate iUTX−/− mice with a full body tamoxifen-inducible deletion of UTX de novo, B6.129-Gt(ROSA)26Sor^tm1(cre/ERT2)Tyj/J (Rosa26ERT2Cre) mice were crossed to Kdm6afl/fl mice to generate F2 progeny Rosa26ERT2Cre/+Kdm6afl/fl offspring. CD451x2 mice were generated de novo by crossing CD45.1+ B6.SJL and CD45.2+ C57BL/6 mice. For all mouse experiments, 6- to 8-week-old age-matched littermates were used in accordance with approved institutional protocols. mBMC mice were generated by depleting host CD451x2 mice by intraperitoneal (i.p.) injection of busulfan (1 mg ml−1) at 20 mg per kg body weight for 3 consecutive days, followed by reconstitution 24 h later with various mixtures (specified for each experiment in figure legends) of bone marrow cells from WT (CD45.1+) and knockout (CD45.2+) donor mice in the presence of anti-NK1.1 (1 mg ml−1; clone PK136) to deplete any remaining mature NK cells. For inducible deletion of UTX in iUTX−/− mice, mBMC mice produced with WT and iUTX−/− bone marrow were treated with 1 mg tamoxifen i.p. daily for 3 d before MCMV infection. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications63. Experimental cohorts were randomized within cages to avoid cage-specific effects. Littermate cre-negative WT controls for experimental mice were used whenever possible.
MCMV infection
MCMV (Smith strain)64 was serially passaged through BALB/c hosts three times, and then salivary gland viral stocks were prepared with a dounce homogenizer for dissociating the salivary glands of infected mice 3 weeks after infection. Experimental mice in studies were infected with MCMV by i.p. injection of 7.5 × 103 plaque-forming units in 0.5 ml of PBS. Mice were monitored and weighed daily and killed when body weight dropped over 20% from initial weight. A ‘sublethal dose’ of MCMV was determined by infection of mice with different dosages to identify the dose in which WT C57BL/6 would survive after 7 d.
Isolation and enrichment of mouse NK cells
Mouse spleens, livers, lungs and blood were collected and prepared into single-cell suspensions as described previously65. Splenic single-cell suspensions were lysed in red blood cell lysis buffer and resuspended in EasySep buffer (StemCell). To avoid depleting Ly6C+ NK cells, we developed a custom antibody cocktail as follows: splenocytes were labeled with 10 μg per spleen of biotin-conjugated antibodies against CD3 (17A2), CD19 (6D5), CD8 (53–6.7), CD88 (20/70), Ly6G (1A8), SiglecF (S17007L), TCRβ (H57-597), CD20 (SA275A11), CD172a (P84) and magnetically depleted from total splenocyte suspensions with the use of anti-biotin-coupled magnetic beads (BioLegend)30.
Ex vivo stimulation of mouse NK cells
For cytokine stimulation experiments, 2 × 104 mouse NK cells were stimulated for 4 h in CR-10 (RPMI 1640 + 25 mM HEPES (Gibco-15630-080) + 10% FBS, 1% l-glutamine, 1% 200 mM sodium pyruvate, 1% MEM-NEAA, 1% penicillin–streptomycin, 0.5% sodium bicarbonate and 0.01% 55 mM 2-mercaptoethanol), brefeldin A (1:1,000 dilution; BioLegend) and monensin (2 µM; BioLegend) with or without recombinant mouse IL-15 (50 ng ml−1; PeproTech), mouse IL-12 (20 ng ml−1; PeproTech) and/or recombinant mouse IL-18 (10 ng ml−1; BioLegend, 767002). Cells were cultured in CR-10 medium alone as a negative control (no treatment). The absolute number of IFN-γ-producing NK cells were determined by acquiring and counting individual cells with an Attune NxT after intracellular flow cytometry staining to determine cell count of IFN-γ+ of the 2 × 104 cells plated per condition. For plate-bound antibody stimulation experiments, 2 × 104 isolated NK cells for each condition were stimulated with 4 mg ml−1 precoated antibody against NK1.1 (PK136) for 4 h in complete medium containing brefeldin A (1:1,000 dilution; BioLegend) and monensin (2 µM; BioLegend). Cells were cultured in medium alone as a negative control (no treatment).
Proliferation assays
CellTrace CFSE (Thermo) stock solution was prepared according to the manufacturers’ instructions and diluted at 1:10,000 in 37 °C PBS. Isolated NK cells were incubated in 0.5 ml of diluted CFSE solution for 5 min at 37 °C. The solution was quenched with 10× the volume of CR-10 medium. Cells were then washed and plated at 50 ng recombinant mouse IL-15 (PeproTech) and cultured for 4 d to assess proliferation.
Human NK cell culture and stimulation
Human PBMCs from anonymous healthy donors were obtained from leukoreduction filters after platelet apheresis from the UCLA Virology Core. NK cells were isolated using the EasySep Human NK Cell Isolation Kit (Stem Cell Technologies) following the manufacturer’s instructions. Following isolation, cells were maintained in 24-well G-Rex plates (Wilson Wolf) in NK MACS medium (Miltenyi Biotech) supplemented with human IL-2 (100 IU ml−1, PeproTech) and human IL-15 (20 ng ml−1, PeproTech) at a plating density of 5 × 106 cells per well. For cytokine stimulation, either freshly isolated human NK cells or cells activated for 14 d with IL-2/IL-15 were plated with K562 leukemia cells at an effector:target ratio of 2.5:1 in addition to human IL-2 (100 IU ml−1, PeproTech), human IL-15 (20 ng ml−1, PeproTech), human IL-12 (10 ng ml−1, PeproTech) and/or human IL-18 (100 ng ml−1, PeproTech) in CR-10 media. NK cells were stimulated with cytokines for 16 h before analysis by flow cytometry.
Cleaved caspase 3 induction
Around 2 × 104 mouse NK cells were cultured in CR-10 media with 5 ng ml−1 recombinant mouse IL-15 (PeproTech; 210-15) and control DMSO or 2.5 μM Nutlin-3a (MedChem Express, HY-10029) for 24 h before intracellular staining for cleaved caspase 3 by flow cytometry.
Adoptive transfer of NK cells
Freshly isolated congenically distinct NK cells were labeled with CTV (Thermo Fisher, C34557) following the manufacturer’s instructions. CTV-labeled cells were mixed at a 1:1 ratio, resuspended in PBS, and 5 × 105 cells were injected i.v. into WT recipient mice.
Tumor killing assays
Low-passage MHC class I-deficient MC38 tumor cells were labeled with CTV (Thermo Fisher, C34557) following the manufacturer’s instructions. Freshly isolated splenic NK cells were cultured with 1 × 104 β2 M−/− MC38 cells per well at effector:target ratios of 4:1, 2:1 and 1:1 in the presence of 50 ng ml−1 recombinant mouse IL-15 (PeproTech, 210-15) for 16 h. Remaining CTV-positive tumor cells were quantified by flow cytometry using the Attune NxT Acoustic Focusing cytometer and data were analyzed with FlowJo v10.7.2 (TreeStar).
Flow cytometry and cell sorting
Cells were analyzed for cell surface markers using fluorophore-conjugated antibodies (BioLegend, eBioscience). Cell surface staining was performed in FACS Buffer (2% FBS and 2 mM EDTA in PBS) and intracellular staining was performed by fixing and permeabilizing using the eBioscience Foxp3/Transcription Factor kit (00-5523-00) for intranuclear proteins or BD Cytofix/Cytoperm (554714) kits for cytokines. Flow cytometry was performed using the Attune NxT Acoustic Focusing cytometer (Thermo Fisher) and data were analyzed with FlowJo v10.7.2 (TreeStar). Cell surface and intracellular staining was performed using the following fluorophore-conjugated antibodies: CD45.1 (A20), CD45.2 (104), NK1.1 (PK136), TCRβ (H57-597), CD3 (17A2), IFN-γ (XMG1.2), Ly6C (HK1.4), Bcl-2 (BCL/10C4), CD11b (M1/70), CD27 (LG.3A10), granzyme B (GB11), UTX (N2C1, GeneTex), goat anti-rabbit H&L (Abcam, ab6717), Bim (c34c5), Ki67 (16A8), CD107a (1D4B), cleaved caspase 3 (Asp175), human CD56 (TULY56), human CD3 (UCHT1) and human IFN-γ (B27). Isolated splenic NK cells were sorted using Aria-H Cytometer (BD) to >95% purity.
Quantitative PCR
For quantitative PCR experiments, up to 5 × 105 cells were isolated using NK EasySep magnetic Isolation kit (StemCell, 19855) as detailed above. RNA was isolated from cells using the Quick-RNA Micro-prep kit (Zymo, R1051). Then, RNA was quantified using a NanoDrop and up to 1 μg was used to synthesize cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, 4368813). Then, 1 μl of undiluted cDNA was directly used in a Taqman Real-Time PCR assay in a 386-well plate. Each sample was plated at a minimum of three technical replicates per taqman probe. Taqman probes (Ifng: Mm01168134_m1, Csf2: Mm01290062_m1, Kdm6a: Mm00801998_m1, Eif2s3: Mm01236979_g1, Kdm5c: Mm00840032_m1, Kdm6c (Uty): Mm00447710_m1, Ddx3x: Mm04207948_gH) were ordered from Thermo Fisher Taqman probe catalog (4331182). Normalized expression was calculated as follows: (1) β-actin was used as a housekeeping gene and to normalize the amount of each sample for each probe; then (2), each sample was normalized to female WT for each gene.
ELISA
Around 2 × 104 mouse NK cells were stimulated with recombinant mouse IL-12 (20 ng ml−1; PeproTech, 210-12) and recombinant mouse IL-18 (10 ng ml−1; PeproTech) for 4 h before conditioned medium was harvested and stored at −80 °C. Mouse IFN-γ and GM-CSF were detected using Legend Max ELISA kits (BioLegend) following the manufacturer’s instructions.
Western blot
Protein was extracted from enriched primary splenic NK cells using Pierce RIPA buffer (Thermo Fisher) with Halt protease inhibitor cocktail (Thermo Fisher) and protein concentration was quantified using the Pierce BCA Protein Assay kit (Thermo Fisher). Samples were electrophoresed on NuPage Novex 4–12% Bis-Tris Protein Gels, transferred to PVDF membranes, and blocked overnight at 4 °C with 5% wt/vol nonfat milk in 1× TBS and 0.1% Tween-20. Immunoblots were performed using rabbit anti-UTX (1:1,000 dilution; Cell Signaling Rabbit monoclonal antibody, 33510), rabbit anti-β-actin (1:10,000 dilution; Cell Signaling, CST4970) and goat anti-rabbit horseradish peroxidase secondary antibody (1:20,000 dilution; Thermo Fisher, 31466). Proteins were detected using the SuperSignal West Pico PLUS ECL kit (Thermo Fisher) and visualized using the Azure Biosystems c280 imager.
RNA-seq library construction
RNA was isolated from 50,000 sort-purified NK cells per sample using RNeasy Mini kit (Qiagen). RNA quality was verified using High Sensitivity RNA Screen Tape and excluded samples with an RNA Integrity Number equivalent (RINe) < 6.0. RNA-seq libraries were sequenced using an Illumina HiSeq 4000 platform (single end, 50 bp).
ATAC-seq library construction
ATAC-seq libraries were produced by the Applied Genomics, Computation and Translational Core Facility at Cedars Sinai in the following manner: 50,000 cells per sample were lysed to collect nuclei and treated with Tn5 transposase (Illumina) for 30 min at 37 °C with gentle agitation. The DNA was isolated with DNA Clean & Concentrator Kit (Zymo) and PCR amplified and barcoded with NEBNext High-Fidelity PCR Mix (New England Biolabs) and unique dual indexes (Illumina). The ATAC-seq library amplification was confirmed by real-time PCR, and additional barcoding PCR cycles were added as necessary while avoiding overamplification. Amplified ATAC-seq libraries were purified with DNA Clean & Concentrator Kit (Zymo). The purified libraries were quantified with Kapa Library Quant Kit (KAPA Biosystems) and quality assessed on a 4200 TapeStation System (Agilent). The libraries were pooled based on molar concentrations and sequenced on an Illumina HiSeq 4000 platform (paired end, 100 bp).
CUT&Tag library preparation
For anti-UTX CUT&Tag library preparation, nuclei were isolated with cold nuclear extraction buffer (20 mM HEPES, pH 7.9, 10 mM KCl, 0.1% Triton X-100, 20% glycerol, 0.5 mM spermidine in 1× protease inhibitor buffer) and incubated with activated concanavalin A-coated magnetic beads (Polysciences, 86057-3) in PCR strip tubes at room temperature for 10 min. A 1:100 dilution of primary antibody (anti-UTX Cell Signaling Rabbit monoclonal antibody no. 33510 or IgG isotype control: Cell Signaling Technology, 3900S) in antibody buffer (20 mM HEPES pH 7.5; 150 mM NaCl, 0.5 mM spermidine, 1× protease inhibitor cocktail (Roche), 0.05% digitonin, 2 mM EDTA, 0.1% BSA) was added and nuclei were incubated with primary antibodies overnight at 4 °C. The next day, the strip tubes were incubated on a magnetic tube holder and supernatants were discarded. Secondary antibody (guinea pig anti-rabbit IgG; Fisher Scientific, NBP172763) was added diluted at 1:100 in Dig-Wash (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 1× protease inhibitor cocktail, 0.05% digitonin) and nuclei were incubated for 1 h at room temperature. Nuclei were washed four times in Dig-Wash and then incubated with a 1:20 dilution of pAG–Tn5 adaptor complex (EpiCypher) in Dig-300 buffer (1× protease inhibitor cocktail, 20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine) for 1 h at room temperature. To stop tagmentation, 25 μl Dig-300 buffer with 10 μl 1 M MgCl, 7.5 μl 0.5 M EDTA, 2.5 μl 10% SDS and 5 μl 10 mg ml−1 proteinase K was added to each reaction and incubated at 55 °C for 1 h. DNA was extracted by phenol:chloroform:isoamyl alcohol separation. DNA was barcoded and amplified using the following conditions: a PCR mix of 25 μl NEBNext 2× mix, 2 μl each of barcoded forward and reverse 10 μM primers, and 21 μl of extracted DNA was amplified at: 58 °C for 5 min, 72 °C for 5 min, 98 °C for 45 s, 16× 98 °C for 15 s followed by 63 °C for 10 s, 72 °C for 1 min. Amplified DNA libraries were purified by adding a 1.3× volume of KAPA pure SPRI beads (Roche) to each sample and incubating for 10 min at 23 °C. Samples were placed on a magnet and unbound liquid was removed. Beads were rinsed twice with 80% ethanol, and DNA was eluted with 25 μl TE buffer. All individually i7-barcoded libraries were mixed at equimolar proportions for sequencing on an Illumina NovaSeq 6000 sequencer.
Sequencing data analysis
ATAC-seq and anti-UTX CUT&Tag fastq files were trimmed to remove low-quality reads and adaptors using Cutadapt58 (version 2.3). The reads were aligned to the reference mouse genome (mm10) with bowtie2 (version 2.2.9)43. Peak calling was performed with MACS2 (version 2.1.1)66. Peaks/regions identified as UTX-bound (UTX CUT&Tag) and differentially accessible (ATAC-seq) were annotated using the annotatepeaks.pl function from the HOMER analysis package. To determine the distance to the nearest transcription start site (TSS), we used the default settings in annotatepeaks.pl, which utilizes RefSeq TSSs to determine the closest TSS. For genomic annotation, we used the ‘basic annotation’ output provided by the assignGenomeAnnotation program in annotatePeaks.pl. The TSS was defined from −1 kB to +100 bp, and the transcription termination site (TTS) was defined from −100 bp to +1 kB. ‘Basic annotation’ is based on alignments of RefSeq transcripts to the UCSC hosted mouse genome file (mm10). HTseq (version 0.9.1)67 was used to count the number of reads that overlap each peak per sample. The peak counts for ATAC-seq were analyzed with DESeq2 (version 1.24.0)68 to identify differentially accessible genomic regions. Peaks with adjusted P value < 0.05 were considered significantly differentially accessible. UTX CUT&Tag peak counts were trimmed for low counts <50 and WT was compared with UTXNKD as a control for background signal. The peak counts for ATAC-seq and anti-UTX CUT&Tag were visualized with Integrated Genome Browser (version 9.1.8) using mouse genome 2011. RNA-seq analysis was carried out by first checking the quality of the reads using FastQC. Then, they were mapped with HISAT2 (version 2.2.1) to the mouse genome (mm10). The counts for each gene were obtained by HtSeq67. Differential expression analyses were carried out using DESeq2 (version 1.24.0)68 with default parameters. Genes with an adjusted P value < 0.05 were considered significantly differentially expressed. Sequencing depth normalized counts were used to plot the expression values for individual genes.
Fuzzy c-means clustering69 was used for both ATAC-seq and RNA-seq using significant (adjusted P value and FDR < 0.05, log2 fold change ± 0.5) normalized counts generated from DESeq2. Mfuzz package (version 3.14) within R was used to perform this analysis into six clusters with a membership score of >0.5. The differentially accessible ATAC peaks were analyzed using the findMotifsGenome.pl function from HOMER70 (version 4.9.1) of each cluster to identify enriched cis-regulatory motifs of TFs. Pathway analysis of clustered RNA-seq data was performed using g:Profiler71 using the g:GOSt function. Top relevant pathways were selected from KEGG Biological Pathways and Gene Ontology Pathways (Biological Processes and Molecular Function).
DICE expression data analysis
Gene expression data in sorted human NK cells by donor sex were provided by the Database of Immune Cell Expression, expression quantitative trait loci and Epigenomics (DICE) Project56. Expression data were downloaded as transcripts per million per sorted immune cell type and male or female sex. Expression was then normalized to the average of females.
Data collection and statistical analyses
Data collection and analysis were not performed blind to the conditions of the experiments. Data points were only deemed to be outliers in the case of experimental contamination or technical error and excluded from analysis. For graphs, data are shown as mean ± s.e.m., and unless otherwise indicated, statistical differences were evaluated using a student’s t-test. For graphs containing multiple groups, either one-way (one treatment or condition) or two-way (multiple treatments or conditions) ANOVA with Tukey’s correction for multiple comparisons was used as stated. For Kaplan–Meier survival curve, samples were compared using the log rank (Mantel–Cox) test with correction for testing multiple hypotheses. A P value < 0.05 was considered significant. Graphs were produced and statistical analyses were performed using GraphPad Prism and ggplot2 library in R. Spearman correlation on best fit regression line was performed using ggpubr library in R. Data distribution was assumed to be normal, but this was not formally tested.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequencing datasets are accessible from the Gene Expression Omnibus under accession number GSE185065. Source data are provided with this paper. Further information and requests for resources and reagents should be directed and will be fulfilled by the corresponding authors.
References
Wilkinson, N. M., Chen, H. C., Lechner, M. G. & Su, M. A. Sex differences in immunity. Annu Rev. Immunol. 40, 75–94 (2022).
Klein, S. L. & Flanagan, K. L. Sex differences in immune responses. Nat. Rev. Immunol. 16, 626–638 (2016).
Pardue, M.-L. & Wizemann, T. M. Exploring the biological contributions to human health: does sex matter? The National Academies Press https://doi.org/10.17226/10028 (2001).
Gianella, S. et al. Sex differences in CMV replication and HIV persistence during suppressive ART. Open Forum Infect. Dis. 7, ofaa289 (2020).
Takahashi, T. & Iwasaki, A. Sex differences in immune responses. Science 371, 347–348 (2021).
Patin, E. et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 19, 302–314 (2018).
Huang, Z. et al. Effects of sex and aging on the immune cell landscape as assessed by single-cell transcriptomic analysis. Proc. Natl Acad. Sci. USA 118, e2023216118 (2021).
Talebizadeh, Z., Simon, S. D. & Butler, M. G. X chromosome gene expression in human tissues: male and female comparisons. Genomics 88, 675–681 (2006).
Fang, H., Disteche, C. M. & Berletch, J. B. X inactivation and escape: epigenetic and structural features. Front. Cell Dev. Biol. 7, 219 (2019).
Chen, X. et al. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev. Neurobiol. 68, 265–273 (2008).
Smith-Bouvier, D. L. et al. A role for sex chromosome complement in the female bias in autoimmune disease. J. Exp. Med. 205, 1099–1108 (2008).
Souyris, M. et al. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 3, eaap8855 (2018).
Hammer, Q., Ruckert, T. & Romagnani, C. Natural killer cell specificity for viral infections. Nat. Immunol. 19, 800–808 (2018).
Orange, J. S. Natural killer cell deficiency. J. Allergy Clin. Immunol. 132, 515–525 (2013).
Bukowski, J. F., Warner, J. F., Dennert, G. & Welsh, R. M. Adoptive transfer studies demonstrating the antiviral effect of natural killer cells in vivo. J. Exp. Med. 161, 40–52 (1985).
Brown, M. G. et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292, 934–937 (2001).
Welsh, R. M., Brubaker, J. O., Vargas-Cortes, M. & O’Donnell, C. L. Natural killer (NK) cell response to virus infections in mice with severe combined immunodeficiency. The stimulation of NK cells and the NK cell-dependent control of virus infections occur independently of T and B cell function. J. Exp. Med. 173, 1053–1063 (1991).
Bancroft, G. J., Shellam, G. R. & Chalmer, J. E. Genetic influences on the augmentation of natural killer cells during murine cytomegalovirus infection: correlation with patterns of resistance. J. Immunol. 126, 988–994 (1981).
Menees, K. B. et al. Sex- and age-dependent alterations of splenic immune cell profile and NK cell phenotypes and function in C57BL/6J mice. Immun. Ageing 18, 3 (2021).
Mujal, A. M., Delconte, R. B. & Sun, J. C. Natural killer cells: from innate to adaptive features. Annu. Rev. Immunol. 39, 417–447 (2021).
Loh, J., Chu, D. T., O’Guin, A. K., Yokoyama, W. M. & Virgin, H. W. T. Natural killer cells utilize both perforin and gamma interferon to regulate murine cytomegalovirus infection in the spleen and liver. J. Virol. 79, 661–667 (2005).
Orange, J. S., Wang, B., Terhorst, C. & Biron, C. A. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin-12 administration. J. Exp. Med. 182, 1045–1056 (1995).
Nakaya, M., Tachibana, H. & Yamada, K. Effect of estrogens on the interferon-gamma producing cell population of mouse splenocytes. Biosci. Biotechnol. Biochem. 70, 47–53 (2006).
Chiossone, L. et al. Maturation of mouse NK cells is a 4-stage developmental program. Blood 113, 5488–5496 (2009).
Wainer Katsir, K. & Linial, M. Human genes escaping X-inactivation revealed by single-cell expression data. BMC Genomics 20, 201 (2019).
Yang, F., Babak, T., Shendure, J. & Disteche, C. M. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622 (2010).
Berletch, J. B. et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 11, e1005079 (2015).
Arnold, A. P. Four core genotypes and XY* mouse models: update on impact on SABV research. Neurosci. Biobehav. Rev. 119, 1–8 (2020).
Hasegawa, H. et al. Activation of p53 by Nutlin-3a, an antagonist of MDM2, induces apoptosis and cellular senescence in adult T-cell leukemia cells. Leukemia 23, 2090–2101 (2009).
Riggan, L. et al. The transcription factor Fli1 restricts the formation of memory precursor NK cells during viral infection. Nat. Immunol. 23, 556–567 (2022).
Min-Oo, G., Bezman, N. A., Madera, S., Sun, J. C. & Lanier, L. L. Proapoptotic Bim regulates antigen-specific NK cell contraction and the generation of the memory NK cell pool after cytomegalovirus infection. J. Exp. Med. 211, 1289–1296 (2014).
Louis, C. et al. NK cell-derived GM-CSF potentiates inflammatory arthritis and is negatively regulated by CIS. J. Exp. Med. 217, e20191421 (2020).
Bjorkstrom, N. K., Strunz, B. & Ljunggren, H. G. Natural killer cells in antiviral immunity. Nat. Rev. Immunol. 22, 112–123 (2022).
Smyth, M. J. et al. Perforin is a major contributor to NK cell control of tumor metastasis. J. Immunol. 162, 6658–6662 (1999).
Van der Meulen, J., Speleman, F. & Van Vlierberghe, P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics 9, 658–668 (2014).
Wang, S. P. et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308–321 (2017).
Gozdecka, M. et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894 (2018).
Wang, C. et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl Acad. Sci. USA 109, 15324–15329 (2012).
Shih, H. Y. et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 165, 1120–1133 (2016).
Miller, S. A., Mohn, S. E. & Weinmann, A. S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 (2010).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Kupz, A. et al. Contribution of Thy1+ NK cells to protective IFN-gamma production during Salmonella typhimurium infections. Proc. Natl Acad. Sci. USA 110, 2252–2257 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Rapp, M. et al. Core-binding factor beta and Runx transcription factors promote adaptive natural killer cell responses. Sci. Immunol. 2, eaan3796 (2017).
Simonetta, F., Pradier, A. & Roosnek, E. T-bet and eomesodermin in NK cell development, maturation and function. Front. Immunol. 7, 241 (2016).
Presnell, J. S., Schnitzler, C. E. & Browne, W. E. KLF/SP transcription factor family evolution: expansion, diversification and innovation in eukaryotes. Genome Biol. Evol. 7, 2289–2309 (2015).
Kramer, B. et al. Early IFN-alpha signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 54, 2650–2669 (2021).
D’Agostino, P. et al. Sex hormones modulate inflammatory mediators produced by macrophages. Ann. N. Y. Acad. Sci. 876, 426–429 (1999).
Lu, F. X. et al. The strength of B cell immunity in female rhesus macaques is controlled by CD8+ T cells under the influence of ovarian steroid hormones. Clin. Exp. Immunol. 128, 10–20 (2002).
Singh, R. P. & Bischoff, D. S. Sex hormones and gender influence the expression of markers of regulatory T cells in SLE patients. Front. Immunol. 12, 619268 (2021).
Cook, K. D. et al. T follicular helper cell-dependent clearance of a persistent virus infection requires T cell expression of the histone demethylase UTX. Immunity 43, 703–714 (2015).
Beyaz, S. et al. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat. Immunol. 18, 184–195 (2017).
Mitchell, J. E. et al. UTX promotes CD8+ T cell-mediated antiviral defenses but reduces T cell durability. Cell Rep. 35, 108966 (2021).
Schmiedel, B. J. et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715 (2018).
Bosselut, R. Pleiotropic functions of H3K27Me3 demethylases in immune cell differentiation. Trends Immunol. 37, 102–113 (2016).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143 (2017).
Hong, S. et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl Acad. Sci. USA 104, 18439–18444 (2007).
Van Laarhoven, P. M. et al. Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24, 4443–4453 (2015).
Rezvani, K. Adoptive cell therapy using engineered natural killer cells. Bone Marrow Transpl. 54, 785–788 (2019).
Eckelhart, E. et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood 117, 1565–1573 (2011).
Lau, C. M. et al. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 19, 963–972 (2018).
Smith, L. M., McWhorter, A. R., Masters, L. L., Shellam, G. R. & Redwood, A. J. Laboratory strains of murine cytomegalovirus are genetically similar to but phenotypically distinct from wild strains of virus. J. Virol. 82, 6689–6696 (2008).
Weizman, O. E. et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 (2017).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Dembele, D. & Kastner, P. Fuzzy C-means method for clustering microarray data. Bioinformatics 19, 973–980 (2003).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Acknowledgements
We thank members of the T.E.O. and M.A.S. laboratories for helpful discussion. We thank the UCLA Technology Center for Genomics and Bioinformatics for RNA-seq library preparation and the Cedars Sinai Applied Genomics, Computation, and Translational Core Facility for ATAC-seq library preparation. T.E.O. is supported by the National Institutes of Health (NIH; AI145997) and UC CRCC (CRN-20-637105). We thank the blood donors and UCLA/CFAR Virology Core Laboratory for providing human PBMCs for study, funded by UCLA CFAR grant 5P30 AI028697. We thank M. Lechner for use of a BioRender license to produce the schematic in Fig. 6. M.A.S. is supported by the NIH (NS107851, AI143894, DK119445), Department of Defense (USAMRAA PR200530) and National Organization of Rare Diseases. M.I.C. is supported by Ruth L. Kirschstein National Research Service Awards (GM007185 and AI007323), and a Whitcome Fellowship from the Molecular Biology Institute at UCLA. L.R. is supported by the Warsaw fellowship from the MIMG department at UCLA. J.H.L. is supported by the NIH NIAMS (T32AR071307) and the UCLA Medical Scientist Training Program (NIH NIGMS T32GM008042). A.P.A. is supported by the NIH (HD100298).
Author information
Authors and Affiliations
Contributions
M.I.C., L.R., J.H.L., R.Y.T., H.H., A.P.A., T.E.O. and M.A.S. designed the study; M.I.C., J.H.L., R.Y.T., L.R. and S.C. performed the experiments; M.I.C., F.M., B.C. and M.P. performed bioinformatics analysis; M.I.C., M.A.S., J.H.L. and T.E.O. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
T.E.O. is a scientific advisor for Modulus Therapeutics and Xyphos companies that have financial interest in human NK cell-based therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Mihalis Verykokakis, Edith Heard, and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the rest of the Nature Immunology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sex differences in IFN-γ production in response to IL-12/18.
a) Representative dot plots showing gating strategy to identify CD3− TCRβ- NK1.1+ mouse NK cells. b) Representative contour plots, (c) percentage IFN-γ+, and (d) normalized IFN-γ MFI of female and male WT NK cells with cultured no treatment (NT) or IL-12 (20 ng/mL) and IL-18 (10 ng/mL) for 4 hours (n = 14 per group). e) Representative contour plots of CD3−CD56+ female (n = 6) and male (n = 7) human NK cells cultured and stimulated with 10 ng/mL of IL-12 for 16 hours in the presence of K562 cells. f) Representative dot plots of splenic NK cells (CD3−TCRβ- NK1.1+) in gonadectomized female and male mice. g) Representative contour plots of total splenic NK cells isolated from gonadectomized female and male mice and cultured with no treatment (NT) or IL-15 (50 ng/mL) and IL-12 (20 ng/mL) for 4 hours. h) Representative contour plots and i) percentage CD27−CD11b− (DN), CD27+CD11b− (CD27 SP), CD27+CD11b+ (DP), and CD27−CD11b+ (CD11b SP) of total splenic NK cells from female and male mice (n = 7 per group). j) Representative contour plots and k) percentage CD27−CD11b− (DN), CD27+CD11b− (CD27 SP), CD27+CD11b+ (DP), and CD27−CD11b+ (CD11b SP) of total splenic NK cells from gonadectomized mice (n = 6 per group). Data are representative of 2-3 independent experiments. Samples were compared using unpaired two-tailed Student’s t test and data points are presented as individual mice with the mean ± SEM (N.S., Not Significant; **, p < 0.01; ****, p < 0.0001). Specific p-values are as follows: c:[NT = 0.987; IL-12+IL-18 = 0.00125]; d:[NT = 0.4144; IL-12+IL-18 < 0.0001]; i:[DN > 0.999; CD27 SP = 0.9514; DP = 0.8995; CD11b SP = 0.6517]; k:[DN = 0.0951; CD27 SP = 0.225; DP = 0.161; CD11b SP = 0.789].
Extended Data Fig. 2 UTX expression in Four Core Genotypes mice and maturation in UTX mouse models.
a) Relative expression of Kdm6a (encodes the protein UTX) by RT-qPCR in Four Core Genotypes mice, in which male or female gonads present are independent of XX or XY chromosome composition, normalized to female WT expression (XX with ovaries) (XX ovaries: n = 4; XY ovaries: n = 5; XX testes: n = 3; XY testes: n = 5). b) Relative UTX MFI in splenic NK cells isolated from female WT (n = 19), male WT (n = 19), female UTXHet (n = 7), and female UTXNKD (n = 4) mice. c) CD11b and CD27 expression within NK cells isolated from female WT (n = 13), male WT (n = 7), and female UTXHet (n = 8) mice. d) Representative western blot showing protein expression of UTX in NK cells isolated from female WT and UTXNKD mice compared to β-actin loading control. Data are representative of 2-3 independent experiments. Samples were compared using a) unpaired two-tailed Student’s t test, b,c) one-way ANOVA with Tukey’s correction for multiple comparisons. Data points are presented as individual mice with the mean ± SEM (N.S., Not Significant; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). Specific p-values are as follows: a:[Ovaries – XX vs. XY < 0.0001; Testes – XX vs. XY = 0.0016]; b:[F WT vs. M WT = 0.003; F UTXNKD vs. F UTXHet and F WT vs. F UTXNKD < 0.0001; M WT vs. F UTXHet = 0.7435; F UTXHet vs. F WT = 0.0004]; c > 0.999.
Extended Data Fig. 3 UTX regulates NK cell fitness.
a) Representative flow cytometry plots of splenic NK cells from female WT, male WT, and female UTXHet, and female UTXNKD mice. b) Representative contour plots and c) percent of WT(CD45.1+) or UTXNKD(CD45.2+) cells of total splenic T (left) and NK (right) cells in 1:1 WT:UTXNKD mBMC mice (n = 3). d) Percentage of Ki67+ cells in blood of 4:1 WT:UTXNKD mBMCs (n = 28), injection ratio used to normalize WT and UTXNKD NK cell numbers. e) Representative histogram (left) and quantification (right) of CFSE expansion, division and proliferation indices calculated using FlowJo’s Proliferation tool of CFSE-labeled splenic NK cells isolated from WT and UTXNKD mice stimulated ex vivo with IL-15 (50 ng/mL) for 4 days (n = 4). f) Schematic showing adoptive transfer of CTV-labeled congenically distinct WT (CD451x2) and UTXNKD (CD45.2+) NK cells transferred into WT (CD45.1+) recipients at a 1:1 ratio with analysis of CTV dilution and WT:UTXNKD ratio on D7 by flow cytometry. g) Representative histograms showing CTV dilution of congenically distinct WT (CD451x2) and UTXNKD (CD45.2+) NK cells transferred into WT (CD45.1+) recipients before transfer (left) and on day 7 post-transfer (right). h) Representative histograms and i) percentage of cleaved caspase 3+ splenic NK cells from female WT and male WT treated ex vivo with IL-15 (5 ng/mL) and DMSO (Female WT: n = 7; Male WT: n = 8) or 2.5 uM Nutlin-3a (Female WT: n = 3; Male WT: n = 4) for 24 hours. j) Representative histograms and k) percentage of cleaved caspase 3+ splenic NK cells from gonadectomized female and male mice (n = 6) treated ex vivo with IL-15 (5 ng/mL) and DMSO or 2.5uM Nutlin-3a for 24 h. l) Representative histograms of Bcl-2 (left) and Bim (right) of flow cytometry in splenic NK cells from female WT and UTXNKD mice (n = 5). “Ctrl”:unstained flow cytometry controls. Data are representative of 2-3 independent experiments. Samples were compared using unpaired two-tailed Student’s t test with Welch’s correction and data points are presented as individual mice with the mean ± SEM (N.S., Not Significant; *, p < 0.05; **, p < 0.01; ****, p < 0.0001). Specific p-values are as follows: c:[T cells=0.705; NK Cells=0.0202]; d < 0.0001; e:[Expansion=0.0192; Division=0.0253; Proliferation=0.0032]; i:[DMSO = 0.0096; Nutlin-3a = 0.0473]; k:[DMSO = 0.001; Nutlin-3a = 0.0013].
Extended Data Fig. 4 UTX enhances effector function independent of gonadal hormone and maturation.
a) Representative contour plots of total NK cells from female WT, male WT, female UTXHet, and female UTXNKD mice cultured with IL-15 (50 ng/mL) and IL-12 (20 ng/mL) for 4 h. b) Absolute number of IFN-γ+ NK cells from female WT, male WT, and female UTXHet mice stimulated with no treatment (NT) or IL-15 (50 ng/mL) and IL-12 (20 ng/mL) for 4 h (n = 8). c) Specific lysis of MHC Class I deficient MC38 (Target) cells by female WT (n = 4), male WT (n = 5), and gonadectomized female (n = 3) and male (n = 6) NK cells for 16 h at 4:1 effector:target ratio, normalized to lysis by female WT. d) Representative histograms and e) MFI of CD107a (n = 5 per group), granzyme b (GzmB) (Female:n = 5; Male:n = 4), and perforin (n = 5) of female WT and male WT NK cells incubated with IL-15 (50 ng/mL) only or additionally stimulated with plate-bound anti-NK1.1 antibody (PK136). “Ctrl” refers to matched unstained control for flow cytometry. Representative f) contour plots of IFN-γ and g) histogram of GzmB expressing total splenic NK cells on D1.5 post MCMV infection of 4:1 WT:UTXNKD mixed bone marrow chimeras (mBMCs), ratio used to normalize cell numbers between genotypes. h) IFN-γ protein production in UTXNKD compared to WT NK cells within maturation subsets: CD27−CD11b− (DN), CD27+CD11b− (CD27 SP), CD27+CD11b+ (DP), and CD27−CD11b+ (CD11b SP) isolated from 4:1 WT:UTXNKD mBMC 1.5 days post-MCMV infection (n = 6). i) Representative contour plots of total NK cells derived from 1:1 WT:iUTX-/- mBMC mice on day 1.5 post MCMV infection, normalized to WT (n = 6). Data are representative of 2-3 independent experiments. Samples were compared using paired two-tailed Student’s t test and data points are presented as individual mice with the mean ± SEM (N.S., Not Significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). Specific p-values are as follows: b:[F WT vs. M WT = 0.0162; M WT vs. UTXHet = 0.968; F WT vs. F UTXHet = 0.0288]; c:[F WT vs. F Gonadectomy=0.245; F WT vs. M WT = 0.0086; M WT vs. M Gonadectomy=0.998; F Gonadectomy vs. M Gonadectomy=0.0247]; e:[CD107a – NT = 0.54 and NK1.1 = 0.772; GzmB – NT = 0.0004 and NK1.1 = 0.00129; Perforin – NT = 0.0101 and NK1.1 < .0001]; h:[DN = 0.03281; CD27 SP = 0.0186; DP = 0.0231; CD11b SP = 0.0114].
Extended Data Fig. 5 UTY is expressed but not sufficient to compensate for loss of UTX in NK cell homeostasis and effector function.
a) Expression in transcripts per million of KDM6A (encodes the protein UTX) and KDM6C (encodes the protein UTY) using DICE database RNA-seq data on sorted NK cells from human females (n = 36) vs. males (n = 54). b) Relative expression of Kdm6a (encodes the protein UTX) and Kdm6c (encodes the protein UTY) by RT-qPCR in splenic NK cells isolated from female WT, male WT, and gonadectomized female and male mice (n = 6 per group). c) Representative flow cytometry dot plots and quantification of d) frequency and absolute numbers of NK cells in spleen of male WT and UTXNKD mice (n = 8 per group). e) Representative flow cytometry contour plots and quantification of f) percentage IFN-γ + and normalized IFN-γ MFI of total NK cells from male WT vs. male UTXNKD mice with no treatment (NT) or in response to IL-15 (50 ng/mL) and IL-12 (20 ng/mL) stimulation for 4 hours ex vivo, MFI normalized to male WT (n = 8 per group). g) Representative flow cytometry contour plots and quantification of h) percentage IFN-γ+ and normalized IFN-γ MFI of total NK cells from male WT vs. male UTXNKD mice with no treatment (NT) or in response to IL-12 (20 ng/mL) and IL-18 (10 ng/mL) stimulation for 4 hours ex vivo, MFI normalized to male WT (n = 8 per group). Data are representative of 2-3 independent experiments. Samples were compared using paired two-tailed Student’s t test and data points are presented as individual mice with the mean ± SEM (N.S., Not Significant; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). Specific p-values are as follows: a < 0.0001; b:[UTX - Female vs. Male=0.0008; rest<0.0001]; d:[%NK = 0.0003; No. NK = 0.002]; f:[%IFN-γ+- NT = 0.112; 15 + 12 = 0.00133; IFN-γMFI – NT = 0.145; 15 + 12 < 0.0001]; h[%IFN-γ+- NT = 0.112; 12 + 18 = 0.00135; IFN-γMFI – NT = 0.155; 12 + 18 < 0.0001].
Extended Data Fig. 6 Integrative ATAC, RNA, anti-UTX CUT&Tag sequencing analysis reveal concomitant changes in chromatin accessibility and transcription mediated by UTX.
a) Schematic of mBMC generated by 1:4 WT (CD45.1+) and UTXNKD (CD45.2+) bone marrow into a lymphodepleted host (CD451x2). After 6wks of reconstitution, splenic NK cells were sorted for ATAC-seq and RNA-seq library preparation. b) Principal component analysis (PCA) of (left) ATAC-seq and (right) RNA-seq changes in WT and UTXNKD NK cells. c) Volcano plot of differentially expressed genes by RNA-seq plotted by Log2FC of UTXNKD vs. WT (x-axis) and Log2 Mean Expression (y-axis). Dotted lines represent Log2FC cut offs (0.5 and 1). Red dots are genes in NK cell effector and developmental pathways. d) Scatter plot highlighting differentially accessible and expressed genes (FDR and p-value<0.05) colored by fuzzy c-means cluster (see Fig. 6). Mean log2FC of ATAC accessibility peaks (y-axis) and log2FC (x-axis) of RNA-seq transcript levels. Best fit regression line (red) with standard error (light red ribbon) (SEM). Positive correlation calculated by two-tailed Spearman correlation of dataset (R = 0.62, p < 2.2×1016). e) Heatmap displaying expression of cell death genes between WT and UTXNKD NK cells FDR < 0.05, adjusted p-value<0.05, and log2FC > 0.5. f) PCA analysis of anti-UTX CUT&Tag in sort-purified WT and UTXNKD NK cells (n = 3). g) Pathway analysis on UTX-bound genes that are decreased (red) or increased (blue) by expression by RNA-seq using Enrichr (p-value by Fisher’s exact test). h) Log2FC in UTXNKDvs.WT of ATAC accessibility (y-axis) plotted by either decreased (>-0.5Log2FC) (blue) or increased (>+0.5Log2FC) (purple) expression by RNA-seq (x-axis) of the UTX bound genes with significant accessibility and expression differences. i) Correlation plot of Log2FC of UTXNKDvs.WT RNA-seq values compared to corresponding ATAC-seq values for each UTX-bound gene. Linear regression was performed (black line) with the standard error 95% confidence intervals plotted (dotted red lines). Two-tailed Pearson’s correlation was performed (r = 0.5165; p-value<0.0001). j) HOMER motif analysis of ATAC-seq peaks grouped by transcription factor family (top) and transcription factor (bottom). Point size indicates percentage of target sequences featuring motif and red gradient indicates -log10(p-value) of enrichment. k) HOMER Motif analysis performed on UTX-bound peaks. % Target Sequences refers to percent of target motifs identified by the HOMER algorithm out of the background motifs. j,k) Cumulative binomial distribution statistical analysis was performed.
Supplementary information
Supplementary Information
Supplementary Data Table 1
Supplementary Data Tables
Supplementary Data Table 2: Table of the differentially accessible peaks between WT and UTX-NKD NK cells. Descriptive statistics are provided. DAG analysis was performed using a two-sided Wilcoxon rank sum test. Supplementary Data Table 3: Table of the differentially expressed genes between WT and UTX-NKD NK cells. Descriptive statistics are provided. DEG analysis was performed using a two-sided Wilcoxon rank sum test. Supplementary Data Table 4: Table of anti-UTX CUT&Tag peaks in WT and UTX-NKD NK cells.
Source data
Source Data Fig. 1
Statistical source data for all graphs in Fig. 1.
Source Data Fig. 2
Statistical source data for all graphs in Fig. 2.
Source Data Fig. 3
Statistical source data for all graphs in Fig. 3.
Source Data Fig. 4
Statistical source data for all graphs in Fig. 4.
Source Data Fig. 5
Statistical source data for all graphs in Fig. 5.
Source Data Fig. 6
Statistical source data for all graphs in Fig. 6.
Source Data Extended Data Fig. 1
Statistical source data for all graphs in Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Statistical source data for all graphs in Extended Data Fig. 2.
Source Data Extended Data Fig. 2
Unprocessed blots for the protein ladder, UTX, and actin from Extended Data Fig. 2d.
Source Data Extended Data Fig. 3
Statistical source data for all graphs in Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Statistical source data for all graphs in Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data for all graphs in Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data for all graphs in Extended Data Fig. 6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, M.I., Li, J.H., Riggan, L. et al. The X-linked epigenetic regulator UTX controls NK cell-intrinsic sex differences. Nat Immunol 24, 780–791 (2023). https://doi.org/10.1038/s41590-023-01463-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01463-8
This article is cited by
-
imply: improving cell-type deconvolution accuracy using personalized reference profiles
Genome Medicine (2024)
-
XX sex chromosome complement modulates immune responses to heat-killed Streptococcus pneumoniae immunization in a microbiome-dependent manner
Biology of Sex Differences (2024)
-
Immune targeting of HIV-1 reservoir cells: a path to elimination strategies and cure
Nature Reviews Microbiology (2024)
-
The conneXion between sex and immune responses
Nature Reviews Immunology (2024)
-
A randomized controlled clinical trial examining the effects of Cordyceps militaris beverage on the immune response in healthy adults
Scientific Reports (2024)