Abstract
Chronic antigen exposure during viral infection or cancer promotes an exhausted T cell (Tex) state with reduced effector function. However, whether all antigen-specific T cell clones follow the same Tex differentiation trajectory remains unclear. Here, we generate a single-cell multiomic atlas of T cell exhaustion in murine chronic viral infection that redefines Tex phenotypic diversity, including two late-stage Tex subsets with either a terminal exhaustion (Texterm) or a killer cell lectin-like receptor-expressing cytotoxic (TexKLR) phenotype. We use paired single-cell RNA and T cell receptor sequencing to uncover clonal differentiation trajectories of Texterm-biased, TexKLR-biased or divergent clones that acquire both phenotypes. We show that high T cell receptor signaling avidity correlates with Texterm, whereas low avidity correlates with effector-like TexKLR fate. Finally, we identify similar clonal differentiation trajectories in human tumor-infiltrating lymphocytes. These findings reveal clonal heterogeneity in the T cell response to chronic antigen that influences Tex fates and persistence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Source data are provided with this paper. All sequencing data generated in this study are available under GEO accession GSE188670.
Code availability
Custom code used in this study is available at https://github.com/katieyost/LCMV-code-2022.
References
Hashimoto, M. et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu. Rev. Med 69, 301–318 (2018).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019).
Blank, C. U. et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Hudson, W. H. et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058.e1044 (2019).
Zander, R. et al. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51, 1028–1042.e1024 (2019).
Beltra, J. C. et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841.e828 (2020).
He, R. et al. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537, 412–428 (2016).
Wu, T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016).
Raju, S. et al. Identification of a T-bethi quiescent exhausted CD8 T cell subpopulation that can differentiate into TIM3+CX3CR1+ effectors and memory-like Cells. J. Immunol. 206, 2924–2936 (2021).
Utzschneider, D. T. et al. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol. 21, 1256–1266 (2020).
Sandu, I. et al. Landscape of exhausted virus-specific CD8 T cells in chronic LCMV infection. Cell Rep. 32, 108078 (2020).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Yao, C. et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8+ T cells. Nat. Immunol. 22, 370–380 (2021).
Yao, C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Belk, J. A., Daniel, B. & Satpathy, A. T. Epigenetic regulation of T cell exhaustion. Nat. Immunol. 23, 848–860 (2022).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Matloubian, M., Kolhekar, S. R., Somasundaram, T. & Ahmed, R. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J. Virol. 67, 7340–7349 (1993).
Pliner, H. A. et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e858 (2018).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Chen, Y. et al. BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol. 22, 996–1007 (2021).
Stelekati, E. et al. Long-term persistence of exhausted CD8 T cells in chronic infection is regulated by microRNA-155. Cell Rep. 23, 2142–2156 (2018).
Chen, J. et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019).
Liu, X. et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019).
Shan, Q. et al. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18, 931–939 (2017).
Martinez, G. J. et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42, 265–278 (2015).
Chung, H. K., McDonald, B. & Kaech, S. M. The architectural design of CD8+ T cell responses in acute and chronic infection: parallel structures with divergent fates. J. Exp. Med. 218, e20201730 (2021).
Evrard, M. et al. Sphingosine 1-phosphate receptor 5 (S1PR5) regulates the peripheral retention of tissue-resident lymphocytes. J. Exp. Med. 219, e20210116 (2022).
Milner, J. J. et al. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl Acad. Sci. USA 117, 25667–25678 (2020).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Qiu, X. et al. Mapping transcriptomic vector fields of single cells. Cell 185, 690–711.e645 (2022).
Pritykin, Y. et al. A unified atlas of CD8 T cell dysfunctional states in cancer and infection. Mol. Cell 81, 2477–2493.e2410 (2021).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016).
Philip, M. & Schietinger, A. Heterogeneity and fate choice: T cell exhaustion in cancer and chronic infections. Curr. Opin. Immunol. 58, 98–103 (2019).
Stadtmauer, E. A. et al. CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365 (2020).
Chang, J. T., Wherry, E. J. & Goldrath, A. W. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15, 1104–1115 (2014).
Daniels, M. A. & Teixeiro, E. TCR signaling in T cell memory. Front. Immunol. 6, 617 (2015).
Mold, J. E. et al. Divergent clonal differentiation trajectories establish CD8+ memory T cell heterogeneity during acute viral infections in humans. Cell Rep. 35, 109174 (2021).
Good, C. R. et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100.e6026 (2021).
Chou, C. et al. The transcription factor AP4 mediates resolution of chronic viral infection through amplification of germinal center B cell responses. Immunity 45, 570–582 (2016).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021).
Ise, W. et al. CTLA-4 suppresses the pathogenicity of self antigen-specific T cells by cell-intrinsic and cell-extrinsic mechanisms. Nat. Immunol. 11, 129–135 (2010).
Letourneur, F. & Malissen, B. Derivation of a T cell hybridoma variant deprived of functional T cell receptor alpha and beta chain transcripts reveals a nonfunctional alpha-mRNA of BW5147 origin. Eur. J. Immunol. 19, 2269–2274 (1989).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e727 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Acknowledgements
We thank the members of the Satpathy, Egawa and Chang laboratories for stimulating discussions; K. Murphy, Washington University, for providing the NFAT-GFP reporter cells; and C.-S. Hsieh, Washington University, for providing the MSCV-based retroviral plasmid MigCaRCh. This work was supported by the National Institutes of Health (NIH) K08CA230188 (A.T.S.), U01CA260852 (A.T.S.), RM1-HG007735 (H.Y.C.), R01AI130152 (T.E.), R21AI161040 (T.E.), a Career Award for Medical Scientists from the Burroughs Wellcome Fund (A.T.S.), a Technology Impact Award and a Lloyd J. Old STAR Award from the Cancer Research Institute (A.T.S.), an ASH Scholar Award from the American Society of Hematology (A.T.S.), the Parker Institute for Cancer Immunotherapy (E.J.W., H.Y.C. and A.T.S.), a Pew-Stewart Scholars for Cancer Research Award (A.T.S.), a Baxter Foundation Faculty Scholar Award, a Leukemia and Lymphoma Society Scholar Award (T.E.) and the Scleroderma Research Foundation (H.Y.C.). H.Y.C. is an investigator of the Howard Hughes Medical Institute. K.E.Y. was supported by the National Science Foundation Graduate Research Fellowship Program (NSF DGE-1656518) and a Stanford Graduate Fellowship and a NCI Predoctoral to Postdoctoral Fellow Transition Award (NIH F99CA253729). K.H.G. was supported by a Stanford School of Medicine Propel Postdoctoral Scholarship. J.A.B was supported by a Stanford Graduate Fellowship and a National Science Foundation Graduate Research Fellowship under grant number DGE-1656518. C.J.R. was funded by an NIH training grant through the Stanford Immunology Program (5T32AI007290-38). S.H. was supported by an NIH training grant (T32CA009547). J.R.G. was supported by a Cancer Research Institute-Mark Foundation Fellowship and T32CA009140. M.B. was supported by the Stanford Innovative Medicines Accelerator (IMA). The sequencing data were generated with instrumentation purchased with NIH funds: S10OD018220 and 1S10OD021763.
Author information
Authors and Affiliations
Contributions
B.D., K.E.Y. and A.T.S. conceptualized the study. B.D., K.E.Y. and A.T.S. wrote and edited the manuscript, and all authors reviewed and provided comments on the manuscript. B.D., K.E.Y., S.H., K.S., K.J.H.G., X.Y., M.B., C.J.R., Q.S. and Y.Q. performed experiments. K.E.Y., S.L.M. and J.A.B. analyzed data. B.D., K.E.Y., J.R.G., E.J.W., H.Y.C., T.E. and A.T.S. guided experiments and data analysis.
Corresponding author
Ethics declarations
Competing interests
A.T.S. is a founder of Immunai and Cartography Biosciences and receives research funding from Allogene Therapeutics and Merck Research Laboratories. H.Y.C. is a cofounder of Accent Therapeutics, Boundless Bio and Cartography Biosciences and an advisor to 10x Genomics, Arsenal Biosciences and Spring Discovery. K.E.Y. is a consultant for Cartography Biosciences. J.A.B. is a consultant for Immunai. E.J.W. is an advisor for Danger Bio, Marengo, Janssen, NewLimit Inc., Pluto Immunotherapeutics, Related Sciences, Rubius Therapeutics, Synthekine and Surface Oncology. E.J.W. is a founder of Surface Oncology, Danger Bio, and Arsenal Biosciences. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
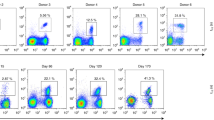
Extended Data Fig. 1 Sorting strategy and quality controls for scATAC-seq data.
(a) Sorting strategy to obtain antigen specific gp33+ and gp33− CD8+ T cells from different organs. (b) Sorting strategy to obtain the main Tex subsets (left). UMAPs of scRNA-seq and scATAC-seq results, originating from the indicated Tex subsets. (c) Bar plot of cell counts from the scRNA-seq samples (top). Stacked bar plot of the phenotypic composition of the indicated scRNA-seq samples (bottom). (d) Quality control of scATAC-seq data. Histogram shows normalized read enrichment on the transcription start sites (TSS) of genes from the indicated samples (top). Density plots depict the cells that passed the TSS enrichment and Log10 unique fragment count threshold. Median TSS enrichment (MTE) is also indicated. (e) Density plots of scATAC-seq data from the main Tex populations depicting the same quality controls as panel (c). (f) UMAP of scATAC-seq data colored by integrated scRNA-seq cluster labels. (g) Heat map of TF motif enrichment at the specific open chromatin regions (OCRs) of the annotated T cell populations (p-values determined by hypergeometric enrichment and adjusted using the Bonferroni correction method).
Extended Data Fig. 2 Characterization of Texint and TexKLR subsets and organ-specific exhaustion signatures.
(a) Ingenuity pathway analyses of the differentially expressed genes identifying enriched biological pathways between the two subsets (Texint versus TexKLR). Top 6 hits are shown. (b) Representative flow cytometry plots that quantify CXCR6 expression in Texterm, Texint and Texprog. Barplots summarize the quantification across three biological replicates. Significant changes were determined by two tailed, unpaired t-test at p < 0.05 (n = 3). Shown are means with SDs. (c) Representative flow cytometry plots show the MKI67+ fractions of the indicated Tex subsets. Boxplot depicts the quantification of MKI67+ Tex subsets. Significant changes were determined by two tailed, unpaired t-test at p < 0.05 (n = 5 biologically independent animals). Box center line, mean; limits, upper and lower quartiles; whiskers, minimum and maximum values. (d) Volcano plots of differentially expressed genes comparing Texterm populations from different organs (log2 FC > 0.25, Bonferroni adjusted p-value < 0.05, p-values determined by two-sided Wilcoxon Rank Sum test). Ingenuity pathway analysis results on the differentially expressed gene groups (bottom). Top 3 hits are shown. (e) Violin plots of the Cell Cycle score of the indicated T cell populations across organs (n = number of scRNA-seq profiles, box center line, median; box limits, upper and lower quartiles; box whiskers, 1.5× interquartile range). P-values determined by two-sided Wilcoxon Rank Sum test relative to overall distribution of single cells from the indicated Tex subsets across all organs. (f) Representative flow cytometry of the MKI67+ fraction of Texterm subsets in the indicated organs. Bar plot summarizes MKI67+ fractions across organs. Significant changes were determined by two tailed, unpaired t-test at p < 0.05 (n = 3 biologically independent animals). Shown are means with SDs.
Extended Data Fig. 3 Analysis of highly-expanded T cell clones in Arm and Cl13 infection.
(a) UMAPs of highly expanded clones from the Arm infection model at the indicated time points. (b) UMAPs of highly expanded clones of the Cl13 infection model at the indicated time points. (c) Stacked bar plot of the phenotypic composition of individual T cell clones with a bias towards the TexKLR fate that also contain some cells with the Texterm phenotype. Top 6 clones are shown. (d) Upset plot of the phenotype combinations of the observed and shuffled TCR clones. Bar represents the mean and error bars represent standard deviation for 50 randomized TCR shuffling iterations performed to obtain the shuffled distribution.
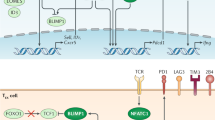
Extended Data Fig. 4 RNA velocity analysis of a divergent clone and regulatory programs of exhaustion.
(a) Heat maps depict the gene expression program of a gp33–reactive divergent clone, differentiating from the Texint stage into either Texterm or TexKLR fates (left). Pseudotime order (direction of differentiation) was determined by RNA velocity analysis and is presented on a UMAP (right). (b) UMAPs of gp33+ CD8+ T cells from Arm infection at D8 and D21 and gp33− at D21. Color gradient (RNA velocity pseudotime order) indicates directions of T cell differentiation fates determined by RNA velocity analysis. (c) UMAP of scATAC-seq results of D8 and D21 gp33+ and gp33−T cells from Cl13 infection. UMAP is colored by the annotated T cell subsets. Small UMAPs (right) show T cells that originate from the indicated gp33 fractions and timepoints. (d) Heat map of Peak score values at the unique open chromatin regions (OCRs) of the T cell subsets determined by scATAC-seq with a list of annotated putative target genes based on proximity (left, log2 FC > 1, FDR < 0.05, p-values determined by two-sided Wilcoxon Rank Sum test and adjusted using the Benjamini & Hochberg procedure to obtain FDRs). Heat map of motif enrichment results at the unique OCR sets of the annotated T cell subsets (right, p-values determined by hypergeometric enrichment and adjusted using the Bonferroni correction method). (e) Upset plot of differentially accessible OCRs relative to Tnaive at the Tox gene locus and their overlap among the different Tex subsets (log2 FC > 1, FDR < 0.01, p-values determined by two-sided Wilcoxon Rank Sum test and adjusted using the Benjamini & Hochberg procedure to obtain FDRs). Violin plot shows the gene expression level of Tox in the identified Tex subsets. Box center line, median; box limits, upper and lower quartiles; box whiskers, 1.5× interquartile range.
Extended Data Fig. 5 Molecular programs of early effector- and progenitor-exhausted T cells and fate mapping experiments.
(a) UMAPs of scATAC-seq (left) and scRNA-seq (right) results from the infection models. Small UMAPs are colored by sample of origin (bottom). (b) Stacked bar plot depicts the phenotypic distribution of gp33+ CD8 T cells from scRNA-seq. (c) Volcano plot of differentially expressed genes (DEGs) between memory precursor T cells (Tmp) from Arm and the precursor exhausted T cells (Texprec) from Cl13 infections (top left). Differential gene expression analyses were performed as follows: log2 FC > 0.25, Bonferroni adjusted p-value < 0.05, p-values determined by two-sided Wilcoxon Rank Sum test. Ingenuity pathway analyses of the Tmp andTexprec specific gene sets. Volcano plot of the differential open chromatin regions (OCRs) of the Tmp and Texprec populations (top right). Differential OCR analyses were performed as follows: log2 FC > 1, FDR < 0.1, p-values determined by two-sided Wilcoxon Rank Sum test and adjusted using the Benjamini & Hochberg procedure to obtain FDRs. Enriched transcription factor (TF) motifs in specific OCRs of Tmp andTexprec subsets are shown (p-values determined by hypergeometric enrichment and adjusted using the Bonferroni correction method). (d) DEGs between the D8 effector T cells (Teff) from Arm and early effector Tex cells (Texeeff) from Cl13 infections (top left). Same statistical approach was used as in (c). Ingenuity pathway analyses of the Teff and Texeeff specific gene sets (bottom left). Volcano plot depicts the differentially accessible OCRs of Teff and Texeeff populations (right). Same statistical approach was used as in (c). Enriched TF motifs in Teff and Texeeff specific OCR sets (p-values determined by hypergeometric enrichment and adjusted using the Bonferroni correction method). (e) Expression of the indicated genes profiled by scRNA-seq. Box center line, mean; limits, upper and lower quartiles; whiskers, minimum and maximum values. (f) Schematic of the Texeeff adoptive transfer experiment with pre-transfer sorting strategy (top). Representative flow cytometry plots show the analysis of the phenotypic content of recovered T cells. Stacked bar plots show the phenotypic distribution from 3 biologically independent animals (mean % of each subset is shown with SDs). (g) Heat map of differentially expressed TFs in Tex subsets. (h) Upset plot of differentially accessible OCRs of TexKLR and Texterm relative to Texint (log2 FC > 1, FDR < 0.01, p-values determined by two-sided Wilcoxon Rank Sum test and adjusted using the Benjamini & Hochberg procedure to obtain FDRs) and their overlap. (i) Schematic of lineage tracing experiment (left). Gating strategy to analyze tdTomato+ fractions of Tex subsets. Boxplot of the % of tdTomato+ Tex subsets are shown. Box center line, mean; limits, upper and lower quartiles; whiskers, minimum and maximum values (n = 4 biologically independent animals).
Extended Data Fig. 6 scRNA/TCR-seq reveals T cell clone behaviors in different organs.
(a) Heat map of the fraction overlap between the TCR repertoires of the indicated gp33+ and gp33− CD8+ T cell subsets from different organs. (b) Stacked bar plot of the phenotypic composition of individual clones with divergent behavior across organs. (c) Schematic of the definition of an expanded, organ-shared T cell clone for clone behavior analysis. Only clones that had at least 5 T cells present in each organ were considered. Shared clone numbers across the organs are indicated (left). Table depicting the number of expanded clones that are shared across tissues and their clone behaviors (right). (d) Gene expression analysis of LCMV-gp transcript (left) and Il21 transcript (right) in indicated organs at D22 following Cl13 infection. Significant changes were determined by two tailed, unpaired t-test (n = 10 biologically independent animals). Box center line, mean; limits, upper and lower quartiles; whiskers, minimum and maximum values.
Extended Data Fig. 7 scRNA-seq reveals the phenotypic composition of gp33−, gp33int, and gp33high T cell subsets.
(a) UMAPs of cells profiled by scRNA-seq in the three gp33 T cell fractions colored by the annotated T cell subsets. (b) Venn diagram shows the overlap of all detected TCR clones among the three gp33 T cell fractions. (c) Heat map depicting TCR repertoire overlap (Morisita index) among the different gp33 fractions from the indicated samples. (d) Pie chart representation of the fraction of the detected clone sizes in the three gp33 T cell fractions. (e) Stacked bar plot of the phenotypic distribution of the unique clones from the three gp33 T cell fractions. (f) UMAPs of unique TCR clones determined by scRNA/TCR-seq and colored by the phenotypic distribution of the three gp33 fractions of T cells. (g) Representative flow cytometry plots depict the gating strategy to analyze the fractions of Tex subsets in gp33−, gp33int, and gp33high CD8+ T cells. Bar plots quantify the frequencies of the indicated Tex subsets in each gp33 fraction. Significant changes were determined by two tailed, unpaired t-test at p < 0.05 (n = 3 biologically independent animals). Shown are means with SDs. (h) Representative flow cytometry plots depict the gating strategy to analyze the functionality (IFNG/LAMP1 double positive CD8+ T cells) of gp33−, gp33int, and gp33high CD8+ T cells. Boxplot quantifies the double positive fractions of T cells in each gp33 fraction. Significant changes were determined by two tailed, unpaired t-test at p < 0.05 (n = 7 biologically independent animals). Box center line, mean; limits, upper and lower quartiles; whiskers, minimum and maximum values. (i) Stacked bar plots show the phenotypic content of TCR clones that were tested for TCR signaling avidity. CDR3 amino acid sequences are indicated.
Extended Data Fig. 8 Human expanded TILs exhibit divergent, Texterm- and TexKLR-biased clone behaviors.
(a) Upset plot depicting the combination of phenotypes (clone behaviors) for expanded TIL clones. For clarity, the top 10 most common clone behaviors are shown. Bar plot shows the number of clones with the indicated phenotypes. (b) UMAPs of representative expanded TIL clones with the indicated clone behaviors.
Supplementary information
Supplementary Table
Supplementary Tables 1–12.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Daniel, B., Yost, K.E., Hsiung, S. et al. Divergent clonal differentiation trajectories of T cell exhaustion. Nat Immunol 23, 1614–1627 (2022). https://doi.org/10.1038/s41590-022-01337-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-022-01337-5