Abstract
Foxp3+ regulatory T (Treg) cells expressing the interleukin (IL)-33 receptor ST2 mediate tissue repair in response to IL-33. Whether Treg cells also respond to the alarmin IL-33 to regulate specific aspects of the immune response is not known. Here we describe an unexpected function of ST2+ Treg cells in suppressing the innate immune response in the lung to environmental allergens without altering the adaptive immune response. Following allergen exposure, ST2+ Treg cells were activated by IL-33 to suppress IL-17-producing γδ T cells. ST2 signaling in Treg cells induced Ebi3, a component of the heterodimeric cytokine IL-35 that was required for Treg cell-mediated suppression of γδ T cells. This response resulted in fewer eosinophil-attracting chemokines and reduced eosinophil recruitment into the lung, which was beneficial to the host in reducing allergen-induced inflammation. Thus, we define a fundamental role for ST2+ Treg cells in the lung as a negative regulator of the early innate γδ T cell response to mucosal injury.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data generated or analyzed to support the findings of this study are available from the corresponding author upon request without restrictions. Source data are provided with this paper.
References
Brunkow, M. E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001).
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Campbell, D. J. & Koch, M. A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 (2011).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Josefowicz, S. Z., Lu, L. F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Panduro, M., Benoist, C. & Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 34, 609–633 (2016).
Curotto de Lafaille, M. A. & Lafaille, J. J. Natural and adaptive Foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 (2009).
Fang, D. & Zhu, J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 214, 1861–1876 (2017).
Koch, M. A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009).
Sefik, E. et al. Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Levine, A. G. et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017).
Kuswanto, W. et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity 44, 355–367 (2016).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T cell function in the intestine. Nature 513, 564–568 (2014).
Liew, F. Y., Girard, J. P. & Turnquist, H. R. Interleukin-33 in health and disease. Nat. Rev. Immunol. 16, 676–689 (2016).
Martin, N. T. & Martin, M. U. Interleukin-33 is a guardian of barriers and a local alarmin. Nat. Immunol. 17, 122–131 (2016).
Guo, L. et al. Innate immunological function of TH2 cells in vivo. Nat. Immunol. 16, 1051–1059 (2015).
Molofsky, A. B., Savage, A. K. & Locksley, R. M. Interleukin-33 in tissue homeostasis, injury and inflammation. Immunity 42, 1005–1019 (2015).
Cayrol, C. et al. Environmental allergens induce allergic inflammation through proteolytic maturation of IL-33. Nat. Immunol. 19, 375–385 (2018).
Delacher, M. et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat. Immunol. 18, 1160–1172 (2017).
Galkina, E. et al. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J. Clin. Invest. 115, 3473–3483 (2005).
Van Dyken, S. J. et al. A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17, 1381–1387 (2016).
Hondowicz, B. D. et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity 44, 155–166 (2016).
Guo, L. et al. IL-1 family members and STAT activators induce cytokine production by TH2, TH17 and TH1 cells. Proc. Natl Acad. Sci. USA 106, 13463–13468 (2009).
Kuperman, D. A. et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 8, 885–889 (2002).
Valladao, A. C., Frevert, C. W., Koch, L. K., Campbell, D. J. & Ziegler, S. F. STAT6 regulates the development of eosinophilic versus neutrophilic asthma in response to Alternaria alternata. J. Immunol. 197, 4541–4551 (2016).
Collison, L. W. et al. The inhibitory cytokine IL-35 contributes to regulatory T cell function. Nature 450, 566–569 (2007).
Vignali, D. A. & Kuchroo, V. K. IL-12 family cytokines: immunological playmakers. Nat. Immunol. 13, 722–728 (2012).
Whitehead, G. S. et al. IL-35 production by inducible co-stimulator-positive regulatory T cells reverses established IL-17-dependent allergic airways disease. J. Allergy Clin. Immunol. 129, 207–215 (2012).
Saenz, S. A., Taylor, B. C. & Artis, D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol. Rev. 226, 172–190 (2008).
Ohnmacht, C. et al. Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993 (2015).
Thornton, A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010).
Weiss, J. M. et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209, 1723–1742 (2012).
Halim, T. Y. et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 40, 425–435 (2014).
Chesne, J. et al. IL-17 in severe asthma. Where do we stand? Am. J. Respir. Crit. Care Med. 190, 1094–1101 (2014).
Nakae, S. et al. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity 17, 375–387 (2002).
Al-Ramli, W. et al. TH17-associated cytokines (IL-17A and IL-17F) in severe asthma. J. Allergy Clin. Immunol. 123, 1185–1187 (2009).
Zuany-Amorim, C. et al. Requirement for γδ T cells in allergic airway inflammation. Science 280, 1265–1267 (1998).
Schnyder-Candrian, S. et al. Interleukin-17 is a negative regulator of established allergic asthma. J. Exp. Med. 203, 2715–2725 (2006).
Hellings, P. W. et al. Interleukin-17 orchestrates the granulocyte influx into airways after allergen inhalation in a mouse model of allergic asthma. Am. J. Respir. Cell Mol. Biol. 28, 42–50 (2003).
Chenuet, P. et al. Neutralization of either IL-17A or IL-17F is sufficient to inhibit house dust mite induced allergic asthma in mice. Clin. Sci. 131, 2533–2548 (2017).
Murdoch, J. R. & Lloyd, C. M. Resolution of allergic airway inflammation and airway hyperreactivity is mediated by IL-17-producing γδ T cells. Am. J. Respir. Crit. Care Med. 182, 464–476 (2010).
Fulkerson, P. C. et al. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc. Natl Acad. Sci. USA 103, 16418–16423 (2006).
Raredon, M. S. B. et al. Single-cell connectomic analysis of adult mammalian lungs. Sci. Adv. 5, eaaw3851 (2019).
Saleh, A., Shan, L., Halayko, A. J., Kung, S. & Gounni, A. S. Critical role for STAT3 in IL-17A-mediated CCL11 expression in human airway smooth muscle cells. J. Immunol. 182, 3357–3365 (2009).
Severa, M. et al. The transcriptional repressor BLIMP1 curbs host defenses by suppressing expression of the chemokine CCL8. J. Immunol. 192, 2291–2304 (2014).
Ebert, L. M., Meuter, S. & Moser, B. Homing and function of human skin γδ T cells and NK cells: relevance for tumor surveillance. J. Immunol. 176, 4331–4336 (2006).
Kohlgruber, A. C. et al. γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol. 19, 464–474 (2018).
Guo, X. J. et al. Lung γδ T cells mediate protective responses during neonatal influenza infection that are associated with type 2 immunity. Immunity 49, 531–544 (2018).
Itohara, S. et al. T cell receptor δ gene mutant mice: independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell 72, 337–348 (1993).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Oboki, K. et al. IL-33 is a crucial amplifier of innate rather than acquired immunity. Proc. Natl Acad. Sci. USA 107, 18581–18586 (2010).
Chen, W. Y., Hong, J., Gannon, J., Kakkar, R. & Lee, R. T. Myocardial pressure overload induces systemic inflammation through endothelial cell IL-33. Proc. Natl Acad. Sci. USA 112, 7249–7254 (2015).
Turnis, M. E. et al. Interleukin-35 limits antitumor immunity. Immunity 44, 316–329 (2016).
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014).
Faustino, L. et al. Regulatory T cells migrate to airways via CCR4 and attenuate the severity of airway allergic inflammation. J. Immunol. 190, 2614–2621 (2013).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Legoux, F. P. & Moon, J. J. Peptide:MHC tetramer-based enrichment of epitope-specific T cells. J. Vis. Exp. https://doi.org/10.3791/4420 (2012).
Lilly, C. M. et al. Eotaxin expression after segmental allergen challenge in subjects with atopic asthma. Am. J. Respir. Crit. Care Med. 163, 1669–1675 (2001).
Thomas, S. Y., Banerji, A., Medoff, B. D., Lilly, C. M. & Luster, A. D. Multiple chemokine receptors, including CCR6 and CXCR3, regulate antigen-induced T cell homing to the human asthmatic airway. J. Immunol. 179, 1901–1912 (2007).
Cho, J. L. et al. Allergic asthma is distinguished by sensitivity of allergen-specific CD4+ T cells and airway structural cells to type 2 inflammation. Sci. Transl. Med. 8, 359ra132 (2016).
Acknowledgements
We thank R. Lee for providing Il1rl1fl/fl mice and L. Wu for providing Il33–/– mice. This work was supported by grants from the National Institutes of Health to A.D.L. (R01AI040618, U19AI095261 and T32HL116275), J.W.G. (K08AI125816), R.A.R. (K08 HL140173) and D.A.A.V. (R01CA203689) and from the National Council of Scientific Development and Technology to L.D.F. (CNPq; 237062/2012-7).
Author information
Authors and Affiliations
Contributions
L.D.F. conceived the study, performed the experiments in the HDM and A. alternata model, analyzed the human samples and wrote the manuscript; J.W.G. performed the experiments in the influenza model and assisted in the interpretation of the experiments in the HDM model; R.A.R. performed the parabiosis experiments; K.N. assisted in the execution of the experiments; D.L.H. recruited the human participants; J.L.C. and B.D.M. performed the SAC studies; J.J.M. provided reagents and expertise for experiments using the HDM-specific tetramer; D.A.A.V. provided the reagents and expertise for experiments studying IL-35; and A.D.L. conceived the study, reviewed and assisted in interpretation of the data and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
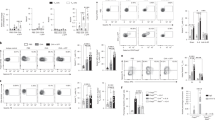
Extended Data Fig. 1 Profiling ST2-expressing Treg and TH cells in the lungs of HDM-treated mice.
a–d, Foxp3YFPcre mice were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 14. a, Schematic of experimental design. b, NanoString analysis of gene expression comparison between flow sort-purified CD4+Foxp3– T helper (TH) cells expressing ST2 (TH2) or not (ST2– TH) from lung parenchyma. Data are presented as volcano plot from three independent experiments of pooled mice (n = 7 mice per experiment). Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used to obtain the P values. c, Representative flow cytometry plots for IL-13 and IL-5 (top), and for IL-17A and IFN-γ (bottom) in TH2 and ST2– TH cells from lung parenchyma. d, Representative histograms showing the expression of the indicated transcription factors in ST2+ Treg cells (red) or ST2– Treg cells (blue) from lung parenchyma. Isotype control is shown in gray.
Extended Data Fig. 2 Subject characteristics.
Aeroallergen-allergic human subjects screened for eligibility with a full medical history, baseline spirometry, methacholine challenge, and allergen skin testing to confirm allergy to either cat dander or Dermatophagoides pteronyssinus (DP). The threshold level of allergen sensitivity was determined by skin prick test titration using serial threefold dilutions of allergen extract.
Extended Data Fig. 3 IL-33 activates ST2+ Treg cells in the lung after HDM exposure.
a–c, Wild-type (WT) and Il33–/– mice were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 10. a, Representative flow cytometry for Foxp3 and ST2 in CD4+ T cells from lung parenchyma. b, Percentage of ST2+ Treg cells, ST2– Treg cells, and TH2 (Foxp3–ST2+) cells in the lung. c, Representative histograms showing the expression of the indicated surface markers in ST2+ Treg and TH2 cells from the lung parenchyma of WT (black) and Il33–/– (gray) mice. Data represent one experiment with n = 6 mice per group of two independent experiments in panels (a–c). Unpaired two-tailed t test was used for statistical analysis in panel (b). Error bars denote mean ± s.d. P values are indicated in the figure.
Extended Data Fig. 4 Treg cell-specific deletion of ST2 does not result in systemic alterations in the number or activation of T cells.
Thymus, spleen, cervical lymph nodes (cLN) and mesenteric lymph nodes (MLN) were harvested from naive Il1rl1fl/flFoxp3YFPcre mice, Il1rl1fl/+Foxp3YFPcre mice, and Il1rl1+/+Foxp3YFPcre (Foxp3YFPcre) littermate control mice for flow cytometric analysis. a, Representative flow cytometry for Foxp3 and CD4 in CD4+ T cells (top) and percentage of Foxp3+ Treg cells (bottom) from the indicated tissues. b, Representative flow cytometry for CD8 and CD4 in CD3+ T cells (top) and percentage of CD8+ T cells and CD4+ T cells (bottom) from the indicated tissues. c, Representative flow cytometry for CD44 and CD62L in CD4+ T cells (top) and percentage of CD44+CD4+ T cells (bottom) from the indicated tissues. Data represent one experiment (Foxp3YFPcre n = 5; Il1rl1fl/+Foxp3YFPcre n = 3; Il1rl1fl/flFoxp3YFPcre n = 4 mice per group) of two independent experiments in panels (a–c). Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis in panels (a–c bottom). No statistical difference was found. Error bars denote mean ± s.d.
Extended Data Fig. 5 Immune response in the lung to HDM in mice with a Treg cell-specific deletion of ST2.
a–c, Il1rl1fl/flFoxp3YFPcre mice and Foxp3YFPcre littermate controls were left untreated (Naive) or were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 14. a, b, Representative flow cytometry for IL-13 and IL-5 (a) and for IFN-γ and IL-17A (b) in CD4+Foxp3– (TH) cells from lung parenchyma. Data represent one experiment with n = 5 mice per group of two independent experiments in panels (a, b). c, Lung explants from Naive and HDM-treated mice were re-stimulated ex vivo with HDM and the indicated cytokines measured in culture supernatants 72 h later by ELISA. Data represent one experiment with n = 4 mice per group of two independent experiments. Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis. Error bars denote mean ± s.d. P values are indicated in the figure.
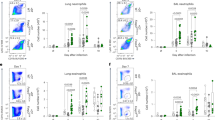
Extended Data Fig. 6 Innate lymphocyte response in the lung to HDM.
a–g, Il1rl1fl/flFoxp3YFPcre mice and Foxp3YFPcre littermate controls were left untreated (Naive) or were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 14. a, Gating strategy for flow cytometric analysis of innate lymphocytes in lung parenchyma. b–d, Percentage of ILC1 (T-bet+), ILC2 (ST2+), and ILC3 (ST2–T-bet–) of CD3–Lin–CD127+ cells (b), percentage of natural killer (NK) cells of CD3–TCRαβ– cells (c), and percentage of NKT cells of CD3+TCRγδ– cells (d) in the lung parenchyma. e–g, Percentage of lung ILCs (e), NK (f), and NKT cells (g) expressing IL-13 and IL-5, IFN-γ, or IL-17A. Data represent one experiment with n = 5 mice per group of three independent experiments in panels (b–g). Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis in panels (b–g). No statistical difference was found. Error bars denote mean ± s.d.
Extended Data Fig. 7 Lung and airway inflammatory response to HDM and A. alternata in mice with a Treg cell-specific deletion of ST2.
a, b, Il1rl1fl/flFoxp3YFPcre mice and Foxp3YFPcre littermate controls were left untreated (Naive) or were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 14. a, Gating strategy for flow cytometric analysis of eosinophils (CD11c–Siglec-F+), neutrophils (Siglec-F–CD11b+Ly6G+), and Ly6C+ inflammatory monocytes (Ly6C+ iMo, Siglec-F–CD11b+Ly6C+) in the lung parenchyma of HDM-treated mice. b, Quantification of cell differential counts in BAL. Mn - Mononuclear cells; Ne - Neutrophils; Eo - Eosinophils. Data represent one experiment (Foxp3YFPcre Naive n = 3; Foxp3YFPcre HDM n = 5; Il1rl1fl/flFoxp3YFPcre Naive n = 3; Il1rl1fl/flFoxp3YFPcre HDM n = 5 mice per group) of two independent experiments. Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis. c, Il1rl1fl/flFoxp3YFPcre mice and Foxp3YFPcre littermate controls were treated with A. alternata i.n. on days 0, 1 and 17-19 and analyzed on day 20. Gating strategy for flow cytometric analysis of eosinophils and neutrophils in the lung parenchyma of A. alternata-treated mice. Error bars denote mean ± s.d. P values are indicated in the figure.
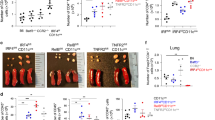
Extended Data Fig. 8 Decreased lung CCL11 and CCL24 and allergic pulmonary inflammation in γδ T cell-deficient mice.
a–c, TCRγδ-deficient (Tcrd–/–) and WT mice were treated with HDM i.n. on days 0 and 7-11 and analyzed on day 14. a, Lung Ccl11 and Ccl24 mRNA levels relative to β2M determined by RT-qPCR. Data represent one experiment with n = 4 mice per group of two independent experiments. b, Number of eosinophils (CD11c–Siglec-F+) and neutrophils (Siglec-F–CD11b+Ly6G+) in lung parenchyma. c, Quantification of cell differential counts in BAL. Mn - Mononuclear cells; Ne - Neutrophils; Eo - Eosinophils. Data represent one experiment (WT n = 5; Tcrd–/– n = 6 mice per group) of two independent experiments in panels (b, c). Unpaired two-tailed t test was used for statistical analysis in panels (a, b) and unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis in panel (c). Error bars denote mean ± s.d. P values are indicated in the figure.
Extended Data Fig. 9 ST2+ Treg cell-derived Ebi3 suppresses γδ T cell responses in the lung to influenza infection.
a–d, Foxp3YFPcre, Il1rl1fl/flFoxp3YFPcre and Ebi3fl/flFoxp3YFPcre mice were infected with PR8 influenza and lungs were harvested for flow cytometry analysis in the indicated time points. a, Representative flow cytometry for IL-17A and IFN-γ in γδ T cells from lung parenchyma on day 11 post-infection. b, Number of IL-17A+ γδ T cells in the lung over the course of infection. Data pooled from two independent experiments (day 0: Foxp3YFPcre n = 5 and Il1rl1fl/flFoxp3YFPcre n = 4; day 4: Foxp3YFPcre n = 18 and Il1rl1fl/flFoxp3YFPcre n = 16; day 7: Foxp3YFPcre n = 10 and Il1rl1fl/flFoxp3YFPcre n = 12; day 11: Foxp3YFPcre n = 9 and Il1rl1fl/flFoxp3YFPcre n = 9 mice from the two experiments in each time point). Unpaired two-tailed t test with Holm-Sidak correction for multiple comparisons was used for statistical analysis in panel (b). c, Representative flow cytometry for TCRαβ and TCRγδ in CD3+ T cells (top) and for IL-17A and IFN-γ in γδ T cells (bottom) from lung parenchyma on day 7 post-infection. d, Number of lung IL-17A+ γδ T cells. Data pooled from two independent experiments (Foxp3YFPcre n = 10; Il1rl1fl/flFoxp3YFPcre n = 8; Ebi3fl/flFoxp3YFPcre n = 10 mice from the two experiments) in panel (d). Ordinary one-way ANOVA with Tukey’s multiple comparisons test was used for statistical analysis in panel (d). Error bars denote mean ± s.d. P values are indicated in the figure.
Supplementary information
Source data
Source Data Fig. 7
Raw NanoString dataset.
Source Data Extended Data Fig. 1
Raw NanoString dataset.
Rights and permissions
About this article
Cite this article
Faustino, L.D., Griffith, J.W., Rahimi, R.A. et al. Interleukin-33 activates regulatory T cells to suppress innate γδ T cell responses in the lung. Nat Immunol 21, 1371–1383 (2020). https://doi.org/10.1038/s41590-020-0785-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0785-3
This article is cited by
-
Interplay of IL-33 and IL-35 Modulates Th2/Th17 Responses in Cigarette Smoke Exposure HDM-Induced Asthma
Inflammation (2024)
-
TCF1–LEF1 co-expression identifies a multipotent progenitor cell (TH2-MPP) across human allergic diseases
Nature Immunology (2024)
-
Endothelial CD34 expression and regulation of immune cell response in-vitro
Scientific Reports (2023)
-
IL-33 promotes double negative T cell survival via the NF-κB pathway
Cell Death & Disease (2023)
-
Regulatory T cell-derived IL-1Ra suppresses the innate response to respiratory viral infection
Nature Immunology (2023)