Abstract
Aire mediates the expression of tissue-specific antigens in thymic epithelial cells to promote tolerance against self-reactive T lymphocytes. However, the mechanism that allows expression of tissue-specific genes at levels that prevent harm is unknown. Here we show that Brg1 generates accessibility at tissue-specific loci to impose central tolerance. We found that Aire has an intrinsic repressive function that restricts chromatin accessibility and opposes Brg1 across the genome. Aire exerted this repressive influence within minutes after recruitment to chromatin and restrained the amplitude of active transcription. Disease-causing mutations that impair Aire-induced activation also impair the protein’s repressive function, which indicates dual roles for Aire. Together, Brg1 and Aire fine-tune the expression of tissue-specific genes at levels that prevent toxicity yet promote immune tolerance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Klein, L., Kyewski, B., Allen, P. M. & Hogquist, K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014).
Takahama, Y., Ohigashi, I., Baik, S. & Anderson, G. Generation of diversity in thymic epithelial cells. Nat. Rev. Immunol. 17, 295–305 (2017).
Derbinski, J., Schulte, A., Kyewski, B. & Klein, L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039 (2001).
Bruserud, Ø., Oftedal, B. E., Wolff, A. B. & Husebye, E. S. AIRE-mutations and autoimmune disease. Curr. Opin. Immunol. 43, 8–15 (2016).
Anderson, M. S. et al. Projection of an immunological self shadow within the thymus by the AIRE protein. Science 298, 1395–1401 (2002).
Long, H. K., Prescott, S. L. & Wysocka, J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016).
Giraud, M. et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. USA 109, 535–540 (2012).
Giraud, M. et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc. Natl. Acad. Sci. USA 111, 1491–1496 (2014).
Yoshida, H. et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl. Acad. Sci. USA 112, E4448–E4457 (2015).
Meredith, M., Zemmour, D., Mathis, D. & Benoist, C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol. 16, 942–949 (2015).
Villaseñor, J., Besse, W., Benoist, C. & Mathis, D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc. Natl. Acad. Sci. USA 105, 15854–15859 (2008).
Jolicoeur, C., Hanahan, D. & Smith, K. M. T-cell tolerance toward a transgenic beta-cell antigen and transcription of endogenous pancreatic genes in thymus. Proc. Natl. Acad. Sci. USA 91, 6707–6711 (1994).
Derbinski, J., Pinto, S., Rösch, S., Hexel, K. & Kyewski, B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl. Acad. Sci. USA 105, 657–662 (2008).
Pinto, S. et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc. Natl. Acad. Sci. USA 110, E3497–E3505 (2013).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Wagner, A., Regev, A. & Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145–1160 (2016).
Sansom, S. N. et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014).
Bansal, K., Yoshida, H., Benoist, C. & Mathis, D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 18, 263–273 (2017).
Brennecke, P. et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol. 16, 933–941 (2015).
Wang, W. et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95, 492–498 (1998).
Bottomley, M. J. et al. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8, 626–633 (2001).
Koh, A. S. et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. USA 105, 15878–15883 (2008).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Manley, N. R. & Condie, B. G. Transcriptional regulation of thymus organogenesis and thymic epithelial cell differentiation. Prog. Mol. Biol. Transl. Sci. 92, 103–120 (2010).
Rossi, S. W. et al. RANK signals from CD4+3– inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204, 1267–1272 (2007).
Senoo, M., Pinto, F., Crum, C. P. & McKeon, F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536 (2007).
Satoh, R. et al. Requirement of Stat3 signaling in the postnatal development of thymic medullary epithelial cells. PLoS Genet. 12, e1005776 (2016).
Lomada, D. et al. Stat3 signaling promotes survival and maintenance of medullary thymic epithelial cells. PLoS Genet. 12, e1005777 (2016).
Natoli, G. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb. Perspect. Biol. 1, a000224 (2009).
Lin, J. et al. Increased generation of Foxp3+ regulatory T cells by manipulating antigen presentation in the thymus. Nat. Commun. 7, 10562 (2016).
Gillard, G. O., Dooley, J., Erickson, M., Peltonen, L. & Farr, A. G. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J. Immunol. 178, 3007–3015 (2007).
Asano, M., Toda, M., Sakaguchi, N. & Sakaguchi, S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J. Exp. Med. 184, 387–396 (1996).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Kadoch, C. et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213–222 (2017).
Miller, E. L. et al. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 24, 344–352 (2017).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Fernández, E. et al. Establishment and characterization of cloned human thymic epithelial cell lines. Analysis of adhesion molecule expression and cytokine production. Blood 83, 3245–3254 (1994).
Murata, S. et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007).
Org, T. et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 (2008).
Koh, A. S., Kingston, R. E., Benoist, C. & Mathis, D. Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc. Natl. Acad. Sci. USA 107, 13016–13021 (2010).
Chakravarty, S., Zeng, L. & Zhou, M. M. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure 17, 670–679 (2009).
Su, M. A. et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 118, 1712–1726 (2008).
Pitkänen, J. et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J. Biol. Chem. 275, 16802–16809 (2000).
Iglesias, P. & Díez, J. J. Management of endocrine disease: a clinical update on tumor-induced hypoglycemia. Eur. J. Endocrinol. 170, R147–R157 (2014).
Schneider, R., Heverhagen, A. E., Moll, R., Bartsch, D. K. & Schlosser, K. Differentiation between thyroidal and ectopic calcitonin secretion in patients with coincidental thyroid nodules and pancreatic tumors—a report of two cases. Exp. Clin. Endocrinol. Diabetes 118, 520–523 (2010).
Falanga, A., Schieppati, F. & Russo, D. Cancer tissue procoagulant mechanisms and the hypercoagulable state of patients with cancer. Semin. Thromb. Hemost. 41, 756–764 (2015).
Stanton, B. Z. et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 49, 282–288 (2017).
Dykhuizen, E. C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Trotter, K. W., King, H. A. & Archer, T. K. Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2β and Ku70/86. Mol. Cell. Biol. 35, 2799–2817 (2015).
Abramson, J., Giraud, M., Benoist, C. & Mathis, D. Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–135 (2010).
Chi, T. H. et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19, 169–182 (2003).
Gordon, J. et al. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7, 69 (2007).
Zhao, Z. et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 24, 1389–1402 (2010).
Gardner, J. M. et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847 (2008).
Jain, R. & Gray, D. H. Isolation of thymic epithelial cells and analysis by flow cytometry. Curr. Protoc. Immunol. 107, 3.26.1–3.26.15 (2014).
Cipolletta, D. et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012).
Su, A. I. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101, 6062–6067 (2004).
Lattin, J. E. et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4, 5 (2008).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Denny, S. K. et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166, 328–342 (2016).
Whyte, W. A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012).
Jia, J. et al. Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell 151, 576–589 (2012).
Morey, L., Aloia, L., Cozzuto, L., Benitah, S. A. & Di Croce, L. RYBP and Cbx7 define specific biological functions of Polycomb complexes in mouse embryonic stem cells. Cell Rep. 3, 60–69 (2013).
Handoko, L. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 (2011).
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 (2006).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
John, S. et al. Genome-scale mapping of DNase I hypersensitivity. Curr. Protoc. Mol. Biol. 103, 21.27.1–21.27.20 (2013).
Anderson, M. S. et al. The cellular mechanism of Aire control of T cell tolerance. Immunity 23, 227–239 (2005).
Acknowledgements
We are grateful to N. Manley (University of Georgia, Athens, GA, USA) for Foxn1ex9Cre mice; N. Hathaway (University of North Carolina, Chapel Hill, NC, USA), C. Kadoch (Harvard Medical School, Boston, MA, USA), S. Braun (Stanford University, Stanford, CA, USA) and E. Chory (Stanford University, Stanford, CA, USA) for CiA constructs; M. Toribio (Universidad Autónoma de Madrid, Madrid, Spain) and D. Mathis (Harvard Medical School, Boston, MA, USA) for the 4D6 cTEC line; D. Mathis, M. Anderson and S. Denny for insightful comments; Y. Chien, C. Weber, J. Kirkland, L. Wagar and J. Ronan for critical reading of the manuscript; and J. Gardner, P. Chu and R. Li for technical assistance. We thank the Stanford Shared FACS facility and S. Kim for flow cytometry and cell sorting. The Stanford BioX3 cluster was used for computational analyses (NIH S10 grant 1S10RR02664701). This work was supported by the Howard Hughes Medical Institute (to G.R.C.), the NIH (grants CA163915 and NS046789 to G.R.C., P50-HG007735 to H.Y.C. and W.J.G., T32HG000044 to J.D.B., and T32 GM007790 to E.L.M.), the Lymphoma and Leukemia Society (A.S.K.) and the National Library of Medicine (Stanford Biomedical Informatics Training Grant LM-07033 to D.M.M.).
Author information
Authors and Affiliations
Contributions
A.S.K. and G.R.C. conceived of the study and wrote the paper. A.S.K. planned and performed all experiments and data analysis. A.S.K., E.L.M., J.D.B. and D.M.M. performed ATAC-seq data analysis. J.W. performed kidney capsule transplants. W.J.G. and H.Y.C. provided conceptual insights and advised on data analysis and experimental design.
Corresponding authors
Ethics declarations
Competing interests
W.J.G. and H.Y.C. are cofounders of Epinomics, Inc. Stanford University has filed a patent on ATAC-seq on which W.J.G. and H.Y.C. are named as inventors.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 mTEChi differentiation promotes major shifts in the chromatin accessibility landscape.
(a) Flow cytometry gating sequence for sorting mTECs. (b) Representative histogram of aggregate Tn5 insertions (blue) and smoothed signal (red) around transcriptional start sites (TSS). (c) Representative distribution of ATAC-seq fragment size exhibiting nucleosomal periodicity. (d) Representative comparison of ATAC-seq fragment density at peaks between biological replicates. (e) Signal enrichment for ATAC-seq libraries, defined by the fold-change between the maximum and minimum signals within the 4 kb region displayed in (b). (f) First principal component (PC) of PCA representing 30.36% of variance separates mTEChi and mTEClo samples. (g) Genomic signal tracks of ATAC-seq fragments at three loci from mTEChi and mTEClo samples. Red arrowheads indicate differentially accessible regions. (h) Representative density plots of ChIP-seq read dyads of indicated factors in embryonic stem cells at ATAC-seq peaks near tissue-specific genes whose accessibilities are induced during mTEChi differentiation. The data shown are from 1 experiment representative of > 20 (a), 16 (b,c), or 4 (d) independent experiments or are from n = 4 (f,g) independent experiments or from pooled data representative of 2 (h) independent experiments.
Supplementary Figure 2 Aire and Brg1 have opposing influences on accessibility.
(a) Representative immunofluorescence stainings of 4-week old thymic sections from indicated genotypes for medullary marker keratin-14 (red) and Aire (green). White scale bar = 10 um. (b) Representative frequencies of TECs in 4-week old thymi from indicated genotypes assessed by flow cytometry. (c) Representative frequencies of mTEChi and mTEClo compartments expressing Aire from 4-week old thymi of indicated mice. (d) CDF plots of indicated accessibility fold-changes at regions classified as differentially accessible in WT vs. Brg1cKO or AireKO mTEChi. P values were determined by Mann-Whitney U-test (two-tailed). (e) Distribution of distances of indicated peak sets to the nearest TSS. (f) Histogram of the distances between indicated peak sets (Fig. 2c) and the nearest TSS. (g) Genomic signal tracks of ATAC-seq fragments at six loci from indicated mTEChi samples (top). ChIP-seq signal tracks from WT mTEChi samples. Red arrowheads indicate differentially accessible regions. (h-j) ATAC-seq analyses on samples generated by Bansal et. al. 2016. (h) First principal component (PC) of PCA separating WT and Aire−/− mTEChi samples. (i) Heatmap of normalized ATAC-seq fragment density at differential peaks (rows). (j) Heatmap of Aire ChIP-seq fragment dyad density at Aire-repressed ATAC-seq peaks. The data shown are from 1 experiment representative of 3 (a), 10 (b,c) or 2 (g) independent experiments or from pooled data representative of 4 (c) or 2 (j) independent experiments or from n = 2 (h,i) independent experiments.
Supplementary Figure 3 Regions in which accessibility is repressed by Aire and induced by Brg1 are enriched for H3K27ac and active topoisomerases.
(a) ChIP-seq fragment dyad density of indicated factors/histones at Aire-induced or Aire-repressed ATAC-seq peaks. (b) Heatmap of ChIP-seq fragment dyad density of indicated factors/histones at Aire-repressed ATAC-seq peaks. (c) ChIP-seq fragment dyad density at Brg1-induced or Brg1-repressed ATAC-seq peaks. The data shown are from pooled data representative of 2 independent experiments (a-c).
Supplementary Figure 4 Aire is dispensable for induced accessibility at tissue-specific loci during mTEC differentiation.
(a) CDF plot of accessibility fold-changes between AireKO mTEChi and AireKO mTEClo at indicated ATAC-seq peaks differentially accessible or unchanged in WT mTEChi vs. mTEClo. (b) CDF plot of transcriptional fold-changes between AireKO mTEChi and AireKO mTEClo at indicated genes upregulated or unchanged by Aire. (c) CDF plot of accessibility fold-changes between WT mTEChi and AireKO mTEChi at indicated ATAC-seq peaks differentially accessible or unchanged in WT mTEChi vs mTEClo. (d) CDF plot of transcriptional fold-changes between WT mTEChi and AireKO mTEChi at indicated genes upregulated or unchanged by Aire. The data shown are from pooled data representative of 2 independent experiments (a-d). P values determined from Mann-Whitney U-tests (two-tailed) (a-d).
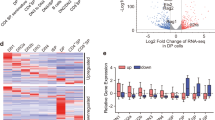
Supplementary Figure 5 Differential enrichment of transcription factor motifs and footprints during mTEC maturation.
(a-c) Change in deviation from expected accessibility signal at ATAC-seq peaks containing known trans-factor motifs (key) between indicated samples. Diamonds represent means, circles represent replicates. (d) NF-kB motifs depicted from analyses in (a-c) and the changes in deviation scores from mTEChi samples of indicated genotypes. Mean +/− s.e.m. (e-p) Differential accessibility footprints in mTEChi vs. mTEClo samples at ATAC-seq peaks containing indicated motifs. The data shown are n = 2 (a-c, d: Aire-KO or Brg1-cKO samples) or n = 4 (d: WT samples) independent experiments, or from pooled data representative of 4 (e-p) independent experiments.
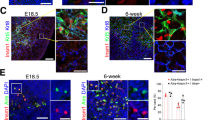
Supplementary Figure 6 T cell compartments in Brg1-cKO mice.
(a,b) Frequencies of thymocyte compartments in 4-week old mice from indicated genotypes assessed by flow cytometry Mean +/− s.e.m. n.s., not significant (two-tailed t-tests). (c) Frequency (left) and cellularity (right) of cTEC compartment in 4-week old mice from indicated genotypes. Mean +/− s.e.m. P values determined by two-tailed t-test < 0.001 (***). (d) Cellularity of thymocyte compartments in 4-week old mice from indicated genotypes. Mean +/− s.e.m. P values determined by two-tailed t-test < 0.05 (*), < 0.01 (**). (e) Cellularity of indicated splenic T cell compartments in 4-week old mice from indicated genotypes. Mean +/− s.e.m. P values determined by two-tailed t-test < 0.01 (**). (f) Frequency (left) and celluarity (right) of FoxP3+CD25+ regulatory T cells (Treg) in spleen from 4 week-old mice of indicated genotypes. Mean +/− s.e.m. P values determined by two-tailed t-test < 0.05 (*). (g) Purified CD4+CD25neg Tconv cells were mixed with CD4+CD25+ Treg from WT and Brg1-cKO mice in a criss-cross fashion as indicated, and assayed for proliferation in the presence of irradiated splenocytes and anti-CD3. Mean +/− s.e.m. (h) Histological analyses of indicated tissues from 6 month-old WT or Brg1-cKO mice via H&E and anti-CD3 immunohistochemistry stainings for infiltrating lymphocytes at indicated peripheral tissues. Scale bars for 10x, 60x images = 200 um, 50 um, respectively. The data shown are from 1 experiment representative of 8 (a) or 2 (h) independent experiments or from n = 8 (b–f) or 3 (g) independent experiments.
Supplementary Figure 7 Aire and BAF have divergent influences on accessibility after recruitment to chromatin.
(a) The CiA:Oct4 locus exhibits DNase I sensitivity compared to the inaccessible rhodopsin (Rho) locus. Mean +/− s.e.m. (b) Rapid maximal reduction in DNase I sensitivity after recruitment of Aire to CiA:Oct4 locus via rapamycin. (c) Schematic of CiA recruitment system. (d) Changes in DNase I sensitivity at CiA:Oct4 locus post BAF vs. Aire recruitment. Mean +/− s.e.m. (e) Recruitment of BAF nor Aire to CiA:Oct4 locus activates transcription. (f) The PSMB11 locus exhibits DNase I sensitivity . Mean +/− s.e.m. (g) Schematic of dCas9 recruitment system: Aire fused to MS2 viral domain is targeted by guide RNA with MS2-binding aptamers. (h) Changes in DNase I sensitivity upon dCas9-induced Aire recruitment to PSMB11 locus, compared to changes at locus encoding ribosomal subunit (RPS29). Mean +/− s.e.m. (i) Model of rheostatic chromatin control of ectopic transcription of tissue-specific genes by Aire and Brg1: BAF chromatin remodeling complexes (orange) coordinate with transcription factors (purple) to poise and recruit transcriptional machinery (gold and gray) to tissue-specific loci during mTEC differentiation. Productive elongation of Pol II is inhibited by negative elongation factors, e.g. Nelf. Subsequent Aire expression and targeting brings interacting positive elongation factors (light blue) to release paused Pol II. Aire’s repressive function inhibits chromatin accessibility, reducing the occupancy of BAF, transcription factors and transcriptional machinery (indicated by fading opacity) and restraining amplitude of transcription. The data shown are from n = 6 (a), 1 (b), or 3 (d,f,h) independent experiments or from 1 experiment representative of 4 (e) independent experiments.
Supplementary information
Supplementary Text, Figures and Table
Supplementary Figures 1–7 and Supplementary Table 1
Rights and permissions
About this article
Cite this article
Koh, A.S., Miller, E.L., Buenrostro, J.D. et al. Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat Immunol 19, 162–172 (2018). https://doi.org/10.1038/s41590-017-0032-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-017-0032-8
This article is cited by
-
Z-DNA marks the spot for AIRE
Cell Research (2024)
-
AIRE relies on Z-DNA to flag gene targets for thymic T cell tolerization
Nature (2024)
-
Insm1 regulates mTEC development and immune tolerance
Cellular & Molecular Immunology (2023)
-
Autoantibodies against type I IFNs in humans with alternative NF-κB pathway deficiency
Nature (2023)
-
Thymic self-antigen expression for immune tolerance and surveillance
Inflammation and Regeneration (2022)