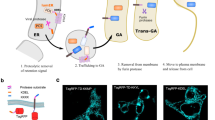
Abstract
Cell-based therapies represent potent enabling technologies in biomedical science. However, current genetic control systems for engineered-cell therapies are predominantly based on the transcription or translation of therapeutic outputs. Here we report a protease-based rapid protein secretion system (PASS) that regulates the secretion of pretranslated proteins retained in the endoplasmic reticulum (ER) owing to an ER-retrieval signal. Upon cleavage by inducible proteases, these proteins are secreted. Three PASS variants (chemPASS, antigenPASS and optoPASS) are developed. With chemPASS, we demonstrate the reversal of hyperglycemia in diabetic mice within minutes via drug-induced insulin secretion. AntigenPASS-equipped cells recognize the tumor antigen and secrete granzyme B and perforin, inducing targeted cell apoptosis. Finally, results from mouse models of diabetes, hypertension and inflammatory pain demonstrate light-induced, optoPASS-mediated therapeutic peptide secretion within minutes, conferring anticipated therapeutic benefits. PASS is a flexible platform for rapid delivery of therapeutic proteins that can facilitate the development and adoption of cell-based precision therapies.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data associated with this study are present in the paper or the Supplementary Information. All genetic components related to this paper are available with a material transfer agreement and can be requested from H.Y. Source data are provided with this paper.
References
Kitada, T., DiAndreth, B., Teague, B. & Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018).
Cubillos-Ruiz, A. et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 20, 941–960 (2021).
Lim, W. A. The emerging era of cell engineering: harnessing the modularity of cells to program complex biological function. Science 378, 848–852 (2022).
Yang, L. et al. Engineering genetic devices for in vivo control of therapeutic T cell activity triggered by the dietary molecule resveratrol. Proc. Natl Acad. Sci. USA 118, e2106612118 (2021).
Yu, Y. et al. Optogenetic-controlled immunotherapeutic designer cells for post-surgical cancer immunotherapy. Nat. Commun. 13, 6357 (2022).
Ye, H. et al. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng. 1, 0005 (2017).
Isabella, V. M. et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018).
Yin, J. et al. A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Sci. Transl. Med. 11, 2318 (2019).
Li, H. S. et al. Multidimensional control of therapeutic human cell function with synthetic gene circuits. Science 378, 1227–1234 (2022).
Mansouri, M. & Fussenegger, M. Therapeutic cell engineering: designing programmable synthetic genetic circuits in mammalian cells. Protein Cell 13, 476–489 (2022).
Wu, C. Y., Roybal, K. T., Puchner, E. M., Onuffer, J. & Lim, W. A. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015).
Wang, Y. et al. A versatile genetic control system in mammalian cells and mice responsive to clinically licensed sodium ferulate. Sci. Adv. 6, eabb9484 (2020).
Stanley, S. A. et al. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336, 604–608 (2012).
Ye, H., Daoud-El Baba, M., Peng, R. W. & Fussenegger, M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568 (2011).
Shao, J. et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci. Transl. Med. 9, eaal2298 (2017).
Zhou, Y. et al. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat. Biotechnol. 40, 262–272 (2022).
Kuwasaki, Y. et al. A red light-responsive photoswitch for deep tissue optogenetics. Nat. Biotechnol. 40, 1672–1679 (2022).
Roybal, K. T. et al. Engineering T cells with customized therapeutic response programs using synthetic notch receptors. Cell 167, 419–432 (2016).
Hyrenius-Wittsten, A. et al. SynNotch CAR circuits enhance solid tumor recognition and promote persistent antitumor activity in mouse models. Sci. Transl. Med. 13, eabd8836 (2021).
Bratanova-Tochkova, T. K. et al. Triggering and augmentation mechanisms, granule pools, and biphasic insulin secretion. Diabetes 51, S83–S90 (2002).
Goodge, K. A. & Hutton, J. C. Translational regulation of proinsulin biosynthesis and proinsulin conversion in the pancreatic β-cell. Semin. Cell Dev. Biol. 11, 235–242 (2000).
Dietz, J. R. Mechanisms of atrial natriuretic peptide secretion from the atrium. Cardiovasc. Res. 68, 8–17 (2005).
Nakagawa, Y., Nishikimi, T. & Kuwahara, K. Atrial and brain natriuretic peptides: hormones secreted from the heart. Peptides 111, 18–25 (2019).
Mellman, I. & Nelson, W. J. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9, 833–845 (2008).
Stockklausner, C., Ludwig, J., Ruppersberg, J. P. & Klöcker, N. A sequence motif responsible for ER export and surface expression of Kir2.0 inward rectifier K+ channels. FEBS Lett. 493, 129–133 (2001).
Kelly, R. B. Pathways of protein secretion in eukaryotes. Science 230, 25–32 (1985).
Zerangue, N. et al. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc. Natl Acad. Sci. USA 98, 2431–2436 (2001).
Sanchez, M. I. & Ting, A. Y. Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods 17, 167–174 (2020).
Wehr, M. C. et al. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods 3, 985–993 (2006).
Liang, F. S., Ho, W. Q. & Crabtree, G. R. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal 4, rs2 (2011).
Foight, G. W. et al. Multi-input chemical control of protein dimerization for programming graded cellular responses. Nat. Biotechnol. 37, 1209–1216 (2019).
Rivera, V. M. et al. A humanized system for pharmacologic control of gene expression. Nat. Med. 2, 1028–1032 (1996).
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Praznik, A. et al. Regulation of protein secretion through chemical regulation of endoplasmic reticulum retention signal cleavage. Nat. Commun. 13, 1323 (2022).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780–791 (2016).
Ramos, C. A. et al. In vivo fate and activity of second- versus third-generation CD19-specific CAR-T cells in B cell non-Hodgkin’s lymphomas. Mol. Ther. 26, 2727–2737 (2018).
Emiliani, V. et al. Optogenetics for light control of biological systems. Nat. Rev. Methods Primers 2, 55 (2022).
Ye, H. & Fussenegger, M. Optogenetic medicine: synthetic therapeutic solutions precision-guided by light. Cold Spring Harb. Perspect. Med. 9, a034371 (2019).
Jung, H. et al. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 10, 314 (2019).
Sun, P., Austin, B. P., Tözsér, J. & Waugh, D. S. Structural determinants of tobacco vein mottling virus protease substrate specificity. Protein Sci. 19, 2240–2251 (2010).
Canini, L. et al. A pharmacokinetic/viral kinetic model to evaluate the treatment effectiveness of danoprevir against chronic HCV. Antivir. Ther. 20, 469–477 (2015).
Therrien, J. P. et al. A gene therapy approach for long-term normalization of blood pressure in hypertensive mice by ANP-secreting human skin grafts. Proc. Natl Acad. Sci. USA 107, 1178–1183 (2010).
Dueñas, M., Ojeda, B., Salazar, A., Mico, J. A. & Failde, I. A review of chronic pain impact on patients, their social environment and the health care system. J. Pain. Res. 9, 457–467 (2016).
Wang, H., Xie, M., Charpin-El Hamri, G., Ye, H. & Fussenegger, M. Treatment of chronic pain by designer cells controlled by spearmint aromatherapy. Nat. Biomed. Eng. 2, 114–123 (2018).
Vlahos, A. E. et al. Protease-controlled secretion and display of intercellular signals. Nat. Commun. 13, 912 (2022).
Mansouri, M., Ray, P. G., Franko, N., Xue, S. & Fussenegger, M. Design of programmable post-translational switch control platform for on-demand protein secretion in mammalian cells. Nucleic Acids Res. 51, e1 (2023).
Stefanov, B. A. et al. Genetically encoded protein thermometer enables precise electrothermal control of transgene expression. Adv. Sci. (Weinh.) 8, e2101813 (2021).
Daringer, N. M., Dudek, R. M., Schwarz, K. A. & Leonard, J. N. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol. 3, 892–902 (2014).
Scheller, L., Strittmatter, T., Fuchs, D., Bojar, D. & Fussenegger, M. Generalized extracellular molecule sensor platform for programming cellular behavior. Nat. Chem. Biol. 14, 723–729 (2018).
Lee, D. et al. Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat. Methods 14, 495–503 (2017).
Saito, H., Kubota, M., Roberts, R. W., Chi, Q. & Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 (2004).
Nagano, K., Ishida, J., Unno, M., Matsukura, T. & Fukamizu, A. Apelin elevates blood pressure in ICR mice with L‑NAME‑induced endothelial dysfunction. Mol. Med. Rep. 7, 1371–1375 (2013).
Chang, M., Smith, S., Thorpe, A., Barratt, M. J. & Karim, F. Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping. Mol. Pain. 6, 56 (2010).
Bonin, R. P., Bories, C. & De Koninck, Y. A simplified up-down method (SUDO) for measuring mechanical nociception in rodents using von Frey filaments. Mol. Pain. 10, 26 (2014).
Acknowledgements
This work was financially supported by grants from the National Key R&D Program of China, Synthetic Biology Research (2019YFA0904500), the National Natural Science Foundation of China (NSFC; 32250010, 32261160373 and 31971346), the Science and Technology Commission of Shanghai Municipality (22N31900300), the Open Research Project of Shanghai Key Laboratory of Diabetes Mellitus (SHKLD-KF-2201) and the Fundamental Research Funds for the Central Universities to H.Y. This work was partially supported by the Science and Technology Commission of Shanghai Municipality (23YF1410700) and China Postdoctoral Science Foundation (2022M721163) and the Natural Science Foundation of Chongqing (CSTB2023NSCQ-MSX0126) to Y.Z. This work was also partially supported by the Science and Technology Commission of Shanghai Municipality (22YF1411400) and China Postdoctoral Science Foundation (2021M701224 and BX20220112) to G.Y. We thank the ECNU Multifunctional Platform for Innovation (011) for supporting the mouse studies and the Instruments Sharing Platform of School of Life Sciences, East China Normal University. We also thank the support from the Chinese Academy of Sciences Youth Interdisciplinary Team. We thank W.D. Heo from the Korea Advanced Institute for Science and Technology (KAIST) for providing pMag and nMag plasmids.
Author information
Authors and Affiliations
Contributions
H.Y. conceived the project. H.Y. and X.W. designed the project. X.W., L.K., D.K., D.D. and X.W. performed the experimental work. H.Y., X.W., L.K., D.K. and Y.Z. analyzed the results and wrote the manuscript. X.W. and L.K. designed, constructed and characterized the performance of chemPASS, antigenPASS and optoPASS. D.K. and G.Y. contributed to the preparation of microcapsule implants. X.W. and Y.Z. helped the animal experiments. D.D. performed flow cytometry experiments. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.Y., X.W. and L.K. are inventors of patent applications (Chinese patent application: 202310413461.0) filed by ECNU that cover the PASS platform. The remaining authors declare no conflicts of interest.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Assessment of the compatibility of ER-retrieved protein.
a, The effect of ER-retrieved protein on the secretion of the continuously expressed Gaussia luciferase. HEK293T cells (6 × 104) were cotransfected with the 50 ng continuously expressed Gaussia luciferase and different amounts of ER-retrieved EGFP plasmid. Gaussia luciferase activity in the culture supernatant was profiled 24 h after transfection. **P = 0.0013 for 50 ng versus 0 ng. b, Viability of HEK293T cells after transfection with various amounts of ER-retrieved EGFP plasmid (0-200 ng). HEK293T cells (1 × 104) were seeded into each well of a 96-well plate and transfected with the indicated amounts of ER-retrieved EGFP plasmid. Twenty-four hours later, cell viability was evaluated with CCK-8 assays. Data are expressed as means ± SD. One-way ANOVA followed by the Dunnett post hoc test was used for multiple comparisons versus the control groups. n = 3 independent experiments (a), n = 6 independent experiments (b). N S, not significant. Detailed descriptions of genetic components and transfection mixtures are provided in Supplementary Tables 1 and 3.
Extended Data Fig. 2 Comparison of different mutated TEVp-mediated secretion of SEAP.
a, Schematic for proteolytic removal of KKRN by different mutated TEVp initiates POI secretion. b, Comparison of different mutated TEVp-mediated secretion of SEAP. HEK293T cells (6 × 104) were cotransfected with ER-loaded SEAP reporter and different plasmids encoding TEVp mutants (pLF14 (S219V), pZQ319 (T30A/S153N/S219V), pZQ326 (I138T/I153N/T180A/S219V). The negative control cells were cotransfected with the same ER-loaded SEAP reporter and the expression vector pcDNA3.1 ( + ). SEAP activity in the culture supernatant was profiled 24 h after transfection. ***P < 0.0001 for all groups versus TEVp -. Data are expressed as means ± SD; n = 3 independent experiments. P values were calculated by One-way ANOVA followed by the Dunnett post hoc test used for multiple comparisons versus the TEVp- group. Detailed descriptions of genetic components and transfection mixtures are provided in Supplementary Tables 1 and 3.
Extended Data Fig. 3 Optimization the amount of DNA used for chemPASS.
a, Optimization of transfection amounts of chemPASSABA plasmids. HEK293T cells (6 × 104) were cotransfected with the 50 ng ER-loaded SEAP reporter and plasmids encoding chemPASSABA (PYL1-cTEVp and ABI-nTEVp) from 100 to 200 ng at a ratio of 1:1 (w/w). At 32 h after transfection, cells were incubated with ABA (100 μM). SEAP activity in the culture supernatant was quantified 24 h after ABA induction. b, Optimization of transfection amounts of chemPASSGZV plasmids. HEK293T cells (6 × 104) were cotransfected with 50 ng ER-loaded SEAP reporter and plasmids encoding chemPASSGZV (nTEVp-NS3a and GNCR1-nTEVp) from 100 to 200 ng at a ratio of 1:1 (w/w). At 32 h after transfection, cells were incubated with GZV (10 μM). SEAP activity in the culture supernatant was quantified 24 h after GZV induction. c, Optimization of transfection amounts of chemPASSDNV plasmids. HEK293T cells (6 × 104) were cotransfected with 50 ng ER-loaded SEAP reporter and plasmids encoding nTEVp-NS3a and DNCR2-cTEVp from 100 to 200 ng at a ratio of 1:1 (w/w). At 32 h after transfection, cells were incubated with DNV (10 μM). SEAP activity in the culture supernatant was quantified 24 h after DNV induction. The optimal transfection amount in (a), (b) and (c) was indicated by red dotted-line boxes. All data are expressed as means ± SD; n = 3 independent experiments. Detailed description of genetic components is provided in Supplementary Table 1.
Extended Data Fig. 4 Quantification of transcriptional controlled protein secretion.
a, Schematic representation of PCA-inducible (PCAON) gene switch. The PCA-inducible gene switch is based on a PCA-triggered transrepressor PcaR (KRAB-PcaV). In the presence of PCA, PcaR is released from PPcaR7 and initiates SEAP expression. e, PCAON switch-mediated SEAP secretion. HEK293T cells (6 × 104) cotransfected with PCAON-encoding plasmids were incubated with PCA (500 μM) at 32 h after transfection. b, Schematic representation of rapamycin-inducible gene switch. In the presence of rapamycin, the transactivator FKBP-VP64 specifically binds to FRB-Gal4 which initiate SEAP expression from a 5 × UAS-specific inducible promoter P5 × UAS (5 × UAS-PhCMVmin). f, Rapamycin-mediated SEAP secretion. HEK293T cells (6 × 104) cotransfected with plasmids encoding FKBP/FRB system were incubated with rapamycin (100 nM) at 32 h after transfection. c, Schematic representation of CRY2/CIBN-inducible gene switch. When exposed to blue light (450 nm), the transactivator (CRY2-VP64) specifically binds to the light sensor domain (CIBN-Gal4) to initiate SEAP expression from a 5 × UAS-specific inducible promoter P5 × UAS (5 × UAS-PhCMVmin). g, CRY2/CIBN system-mediated SEAP secretion. HEK293T cells (6 × 104) were cotransfected with plasmids encoding CRY2/CIBN system and illuminated with blue light (0.5 mW cm−2) for 24 h (alternating 1 min on, 5 min off) or instead kept in the dark at 32 h after transfection. d, Schematic representation of far-red light-inducible gene switch. When exposed to far-red light (730 nm), the bacterial photoreceptor BphS causes intracellular guanylate triphosphate (GTP) conversion into cyclic diguanylate monophosphate (c-di-GMP). The increase in c-di-GMP induces dimerization of the FRL-dependent transcriptional activator p65-VP64-BldD, which further binds to its synthetic promoter PFRL to initiate the SEAP expression. h, Far-red light-controlled transcription device-mediated SEAP secretion. HEK293T cells (6 × 104) cotransfected with plasmids encoding FRL-inducible BphS system were illuminated with far-red light (2 mW cm−2) for 6 h or instead kept in the dark at 32 h after transfection. All data in (e), (f), (g) and (h) are expressed as means ± SD; n = 3 independent experiments. Detailed descriptions of genetic components and transfection mixtures are provided in Supplementary Tables 1 and 3.
Extended Data Fig. 5 Quantification of chemPASS-mediated secretion.
HEK293T cells (6 × 104) cotransfected with plasmids encoding chemPASSDNV were incubated with DNV (1 µM) at 24 h after transfection. SEAP activity in the culture supernatant was profiled at different time points (0-60 h) after DNV induction. ***P < 0.0001 for all comparisons. Data are expressed as means ± SD. n = 3 independent experiments. Student’s t tests. Detailed descriptions of genetic components and transfection mixtures are provided in Supplementary Tables 1 and 3.
Extended Data Fig. 6 Quantification of SEAP secretion driven by two plasmids encoded chemPASSDNV and one plasmid encoded chemPASSDNV.
HEK293T cells (6 × 104) were cotransfected with ER-loaded SEAP reporter and either two plasmids encoding nTEVp-NS3a and DNCR2-cTEVp, or one plasmid encoding nTEVp-NS3a/DNCR2-cTEVp. At 24 h after transfection, cells were incubated with DNV (10 μM). SEAP activity in the culture supernatant was quantified 24 h after DNV induction. ***P < 0.0001 for all comparisons. All data are expressed as means ± SD; n = 3 independent experiments. Student’s t tests. Detailed description of genetic components is provided in Supplementary Tables 1 and 3.
Extended Data Fig. 7 Quantification of antigenPASS-mediated apoptosis by different cell ratios.
a, b, Representative flow cytometry plots using Annexin V-FITC/PI staining for antigenPASS-mediated apoptosis by secreted perforin (a) and granzyme B (b). Jurkat T cells (5 × 104) engineered with an anti-CD19 scFv-synNotch-TEVp receptor were co-cultured with different amounts (5 × 104 or 1.5 × 105) of CD19+ or CD19− HEK293T cells for 24 h and then stained with Annexin V-FITC/PI for flow cytometry analysis. c, d, Percentage of apoptosis of targeted cells in (a) and (b). In c, ***P < 0.0001 for all comparisons in Perforin-early. Apop.; **P = 0.0021 and ***P = 0.0001 for Perforin-late. Apop, left to right. In d, ***P < 0.0001 for all comparisons in Granzyme B-early. Apop., left to right; **P = 0.0045 and ***P = 0.0006 for Granzyme B-late. Apop., left to right. Data are expressed as means ± SD; n = 3 independent experiments. Student’s t tests. Detailed descriptions of genetic components are provided in Supplementary Table 1.
Extended Data Fig. 8 Quantification of antigenPASS, synNotch, and CAR-T cell-mediated secretion of the IL-2.
a, Schematic representation of antigenPASS-mediated IL-2 secretion. Jurkat-T cells were engineered with antigenPASS and ER-loaded IL-2. Stimulation by CD19+ HEK293T sender cells leads to intramembrane proteolysis, allowing TEVp release from its tether and thus get proximity to the ER-loaded, TEVcs-bearing IL-2 which then initiate their secretion by removal of the KKRN retrieval signal. b, Schematic representation of synNotch-mediated IL-2 secretion. Jurkat-T cells were engineered with synNotch system and 5× Gal4 response elements controlling the expression of IL-2. Stimulation by CD19+ HEK293T sender cells lead to intramembrane proteolysis, allowing Gal4-VP64 release from its tether and translocate into the nucleus where it can bind to its synthetic promoter to initiate IL-2 expression. c, Schematic representation of CAR-T-mediated IL-2 secretion. Jurkat-T cells were engineered with anti-CD19−CAR. Stimulation by CD19+ HEK293T sender cells drives Jurkat T cells to produce IL-2. d–f, Jurkat T cells (1 × 105) engineered with antigenPASS (d), SynNoych system (e), or CAR (f) were co-cultured with CD19+ or CD19− HEK293T cells (1 × 105). In d, ***P < 0.0001. In e, ***P < 0.0001. In f, ***P < 0.0001. The IL-2 concentrations in the culture supernatant were quantified by ELISA 24 h after stimulation. All data are expressed as means ± SD; Student’s t test. n = 3 independent experiments. Detailed descriptions of genetic components are provided in Supplementary Table 1.
Extended Data Fig. 9 Quantification of optoPASS-mediated secretion by using CRY2PHR/CIBN and REDMAP systems.
a, Schematic representation of CRY2PHR/CIBN-based optoPASS. Split TEVp fragments were fused to the CRY2PHR and CIBN domains. When exposed to blue light (450 nm), split TEVp quickly recovers its catalytic activity upon light-dependent dimerization of CRY2PHR and CIBN, resulting in protein secretion. b, Screening different combinations of CRY2PHR/CIBN domains and split TEVp fragments. HEK293T cells were transfected with the different combinations (1, CRY2PHR-nTEVp/CIBN-cTEVp; 2, nTEVp- CRY2PHR/CIBN-cTEVp; 3, CRY2PHR-cTEVp/CIBN-nTEVp; 4, cTEVp-CRY2PHR/CIBN-nTEVp; 5, CRY2PHR-nTEVp/cTEVp- CIBN; 6, nTEVp- CRY2PHR/cTEVp-CIBN; 7, CRY2PHR-cTEVp/nTEVp-CIBN; 8, cTEVp-CRY2PHR/nTEVp-CIBN). SEAP activity in the culture supernatant was quantified 24 h after illumination (450 nm, 0.5 mW/cm2). c, Schematic representation of REDMAP-based optoPASS. Split TEVp fragments were fused to the PhyA and FHY1 domains. When exposed to red light (660 nm), split TEVp quickly recovers its catalytic activity upon light-dependent dimerization of ΔPhyA and FHY1, resulting in protein secretion. d, Screening different combinations of ΔPhyA/FHY1 domains and split TEVp fragments. HEK293T cells were transfected with the different combinations (1, ΔPhyA-nTEVp/FHY1-cTEVp; 2, nTEVp-ΔPhyA/FHY1-cTEVp; 3, ΔPhyA-cTEVp/FHY1-nTEVp; 4, cTEVp-ΔPhyA/FHY1-nTEVp; 5, ΔPhyA-nTEVp/cTEVp-FHY1; 6, nTEVp-ΔPhyA/cTEVp-FHY1; 7, ΔPhyA-cTEVp/nTEVp-FHY1; 8, cTEVp-ΔPhyA/nTEVp-FHY1). SEAP activity in the culture supernatant was quantified 24 h after illumination (660 nm, 2 mW/cm2). Data in (b) and (d) are expressed as means ± SD; n = 3 independent experiments.
Extended Data Fig. 10 Optimization of the optoPASS.
a, Schematic representation of the nuclear localization signal (NLS) and nuclear export signal (NES) used in optoPASS. Different light-inducible split TVMVp were generated including, pZQ367 (1, nTVMVp-pMag), pYQ71 (2, NLS-nTVMVp-pMag), pYQ70 (3, NES-nTVMVp-pMag), pZQ365 (4, nMag-cTVMVp), pYQ69 (5, NLS-nMag-cTVMVp), and pYQ68 (6, NES-nMag-cTVMVp). b, Comparison of optoPASS-mediated SEAP secretion with different combinations of NLS and NES. HEK293T cells (6 × 104) were cotransfected with the different combinations shown in (a). c, Schematic depicting the different linkers between nMag/pMag domains and split TVMVp fragments. nMag/pMag were fused to split TVMVp with different linkers including, pYQ68 (L1, NES-nMag-(GGGS)3-cTVMVp), pKL20 (L2, NES-nMag-EAAAK-cTVMVp), pYQ71 (L3, NLS-nTVMVp-(GGGS)3-pMag), and pZQ396 (L4, NLS-nTVMVp-EAAAK-pMag). d, Comparison of the performance of different combinations of linkers of optoPASS. HEK293T cells (6 × 104) were co-transfected with the different combinations shown in (c). e, Schematic depicting the different localization of nTVMVp-pMag domains. The NLS-nTVMVp-pMag fragments are localized in nuclear and the nTVMVp-pMag-KKRN fragments are localized in the ER membrane (through their ER retrieval signal peptides). f, Comparison of the performance of localization of nTVMVp-pMag fragments. The nTVMVp-pMag fusion protein was either free in the cytosol or localized in the ER membrane (dependent on the ER retrieval signal). SEAP activity in the culture supernatant was quantified 4 h after illumination (450 nm, 0.5 mW/cm2). SEAP activity in the culture supernatant was quantified 24 h after illumination (450 nm, 0.5 mW/cm2) for 24 h (alternating 2 min on, 2 min off). All data are expressed as means ± SD; n = 3 independent experiments. Detailed descriptions of genetic components and transfection mixtures are provided in Supplementary Tables 1 and 3.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Supplementary Tables 1–4.
Supplementary Data
Statistical source data for Supplementary Fig. 6.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed immunofluorescence image.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Kang, L., Kong, D. et al. A programmable protease-based protein secretion platform for therapeutic applications. Nat Chem Biol 20, 432–442 (2024). https://doi.org/10.1038/s41589-023-01433-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-023-01433-z
This article is cited by
-
A PASS for protein secretion
Nature Chemical Biology (2024)