Abstract
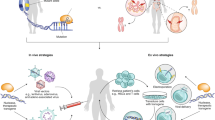
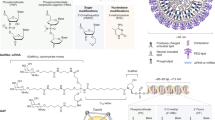
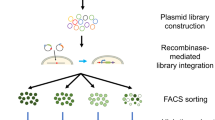
The repertoire of therapeutic proteins has been substantially augmented by molecular engineering approaches, which have seen remarkable advancement in recent years. In particular, advances in directed evolution technologies have empowered the development of custom-designed proteins with novel and disease-relevant functions. Whereas engineered proteins have typically been administered through systemic injection of the purified molecule, exciting progress in gene delivery affords the opportunity to elicit sustained production of the engineered proteins by targeted cells in the host organism. Combining developments at the leading edge of protein engineering and gene delivery has catapulted a new wave of molecular and cellular therapy approaches, which harbor great promise for personalized and precision medicine. This mini-review outlines currently used display platforms for protein evolution and describes recent examples of how the resulting engineered proteins have been incorporated into DNA- and cell-based therapeutic platforms, both in vitro and in vivo. Collectively, the strategies detailed herein provide a framework for synthesizing molecular engineering workflows with gene therapy systems for a breadth of applications in research and medicine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data sharing is not applicable to this paper as no datasets were generated or analyzed during the current study.
References
Ryu DDY, Nam D-H. Recent Progress in Biomolecular Engineering. Biotechnol Prog. 2000;16:2–16.
Mahdavi SZB, Oroojalian F, Eyvazi S, Hejazi M, Baradaran B, Pouladi N, et al. An overview on display systems (phage, bacterial, and yeast display) for production of anticancer antibodies; advantages and disadvantages. Int J Biol Macromol. 2022;208:421–42.
Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–7.
Lee CV, Sidhu SS, Fuh G. Bivalent antibody phage display mimics natural immunoglobulin. J Immunol Methods. 2004;284:119–32.
Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH. Basics of antibody phage display technology. Toxins 2018;10. https://doi.org/10.3390/toxins10060236.
Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM, et al. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front Immunol. 2020;11:1986.
Chasteen L, Ayriss J, Pavlik P, Bradbury ARM. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34:e145.
Hess GT, Cragnolini JJ, Popp MW, Allen MA, Dougan SK, Spooner E, et al. An M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug Chem. 2012;23:1478–87.
Cherf GM, Cochran JR. Applications of yeast surface display for protein engineering. Methods Mol Biol (Clifton, NJ). 2015;1319:155.
Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–7.
Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–44.
Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68.
Valldorf B, Hinz SC, Russo G, Pekar L, Mohr L, Klemm J et al. Antibody display technologies: Selecting the cream of the crop. Biol Chem. 2021. https://doi.org/10.1515/HSZ-2020-0377/ASSET/GRAPHIC/J_HSZ-2020-0377_FIG_005.JPG.
Tsuruta LR, dos ML, Moro AM. Display Technologies for the Selection of Monoclonal Antibodies for Clinical Use. Antibody Eng. 2017. https://doi.org/10.5772/INTECHOPEN.70930.
Zhou C, Jacobsen FW, Cai L, Chen Q, Shen WD. Development of a novel mammalian cell surface antibody display platform. mAbs. 2010;2:508.
Bowers PM, Horlick RA, Kehry MR, Neben TY, Tomlinson GL, Altobell L, et al. Mammalian cell display for the discovery and optimization of antibody therapeutics. Methods. 2014;65:44–56.
Parthiban K, Perera RL, Sattar M, Huang Y, Mayle S, Masters E, et al. A comprehensive search of functional sequence space using large mammalian display libraries created by gene editing. mAbs. 2019;11:884–98.
Kanamori T, Fujino Y, Ueda T. PURE ribosome display and its application in antibody technology. Biochim et Biophys Acta - Proteins Proteom. 2014;1844:1925–32.
He M. Ribosome display: Cell-free protein display technology. Briefings Funct Genom Proteom. 2002;1:204–12.
Kunamneni A, Ogaugwu C, Bradfute S, Durvasula R. Ribosome Display Technology: Applications in Disease Diagnosis and Control. Antibodies. 2020;9:28.
Sockolosky JT, Trotta E, Parisi G, Picton L, Su LL, Le AC, et al. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science. 2018;359:1037–42.
Sedic M, Senn JJ, Lynn A, Laska M, Smith M, Platz SJ, et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet Pathol. 2018;55:341–54.
Haque AA, Dewerth A, Antony JS, Riethmüller J, Latifi N, Yasar H, et al. Modified hCFTR mRNA restores normal lung function in a mouse model of cystic fibrosis. 2017:202853. https://doi.org/10.1101/202853.
Lou B, De Koker S, Lau CYJ, Hennink WE, Mastrobattista E. mRNA Polyplexes with Post-Conjugated GALA Peptides Efficiently Target, Transfect, and Activate Antigen Presenting Cells. Bioconjugate Chem. 2019;30:461–75.
Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol Ther. 2019;27:710–28.
Nault J-C, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–93.
Saunders KO, Wang L, Joyce MG, Yang Z-Y, Balazs AB, Cheng C, et al. Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates from Mucosal Simian-Human Immunodeficiency Virus Infection. J Virol. 2015;89:8334–45.
Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653–8.
Pardi N, Secreto AJ, Shan X, Debonera F, Glover J, Yi Y, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun. 2017;8:14630.
Stadler CR, Bähr-Mahmud H, Celik L, Hebich B, Roth AS, Roth RP, et al. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat Med. 2017;23:815–7.
Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–55.
Mukalel AJ, Riley RS, Zhang R, Mitchell MJ. Nanoparticles for nucleic acid delivery: Applications in cancer immunotherapy. Cancer Lett. 2019;458:102–12.
Malek TR, Yu A, Zhu L, Matsutani T, Adeegbe D, Bayer AL. IL-2 Family of Cytokines in T Regulatory Cell Development and Homeostasis. J Clin Immunol. 2008;28:635–9.
Létourneau S, Krieg C, Pantaleo G, Boyman O. IL-2– and CD25-dependent immunoregulatory mechanisms in the homeostasis of T-cell subsets. J Allergy Clin Immunol. 2009;123:758–62.
Zhang Q, Hresko ME, Picton LK, Su L, Hollander MJ, Nunez-Cruz S, et al. A human orthogonal IL-2 and IL-2Rβ system enhances CAR T cell expansion and antitumor activity in a murine model of leukemia. Sci Transl Med. 2021;13:eabg6986.
Hirai T, Ramos TL, Lin P-Y, Simonetta F, Su LL, Picton LK et al. Selective expansion of regulatory T cells using an orthogonal IL-2/IL-2 receptor system facilitates transplantation tolerance. 2021. https://doi.org/10.1172/JCI139991.
Papadopoulos N, Martin J, Ruan Q, Rafique A, Rosconi MP, Shi E, et al. Binding and neutralization of vascular endothelial growth factor (VEGF) and related ligands by VEGF Trap, ranibizumab and bevacizumab. Angiogenesis. 2012;15:171–85.
Kureshi R, Zhu A, Shen J, Tzeng SY, Astrab LR, Sargunas PR, et al. Structure-Guided Molecular Engineering of a Vascular Endothelial Growth Factor Antagonist to Treat Retinal Diseases. Cel Mol Bioeng. 2020;13:405–18.
June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–5.
Zajc CU, Salzer B, Taft JM, Reddy ST, Lehner M, Traxlmayr MW. Driving CARs with alternative navigation tools – the potential of engineered binding scaffolds. FEBS J. 2021;288:2103–18.
Subklewe M, von Bergwelt-Baildon M, Humpe A. Chimeric Antigen Receptor T Cells: A Race to Revolutionize Cancer Therapy. Transfus Med Hemother. 2019;46:15–24.
Goodman DB, Azimi CS, Kearns K, Garakani K, Garcia J, Patel N et al. Pooled screening of CAR T cells identifies non-native signaling domains for next-generation immunotherapies. Immunology. 2021 https://doi.org/10.1101/2021.07.11.451980.
Lanitis E, Rota G, Kosti P, Ronet C, Spill A, Seijo B, et al. Optimized gene engineering of murine CAR-T cells reveals the beneficial effects of IL-15 coexpression. J Exp Med. 2021;218:e20192203.
Zimmermann K, Kuehle J, Dragon AC, Galla M, Kloth C, Rudek LS, et al. Design and Characterization of an “All-in-One” Lentiviral Vector System Combining Constitutive Anti-GD2 CAR Expression and Inducible Cytokines. Cancers. 2020;12:375.
Chmielewski M, Abken H. CAR T Cells Releasing IL-18 Convert to T-Bethigh FoxO1low Effectors that Exhibit Augmented Activity against Advanced Solid Tumors. Cell Rep. 2017;21:3205–19.
Hu B, Ren J, Luo Y, Keith B, Young RM, Scholler J, et al. Augmentation of Antitumor Immunity by Human and Mouse CAR T Cells Secreting IL-18. Cell Rep. 2017;20:3025–33.
Krenciute G, Prinzing BL, Yi Z, Wu M-F, Liu H, Dotti G, et al. Transgenic Expression of IL15 Improves Antiglioma Activity of IL13Rα2-CAR T Cells but Results in Antigen Loss Variants. Cancer Immunol Res. 2017;5:571–81.
Ataca Atilla P, McKenna MK, Tashiro H, Srinivasan M, Mo F, Watanabe N, et al. Modulating TNFα activity allows transgenic IL15-Expressing CLL-1 CAR T cells to safely eliminate acute myeloid leukemia. J Immunother Cancer. 2020;8:e001229.
Gardner TJ, Lee JP, Bourne CM, Wijewarnasuriya D, Kinarivala N, Kurtz KG, et al. Engineering CAR-T cells to activate small-molecule drugs in situ. Nat Chem Biol. 2022;18:216–25.
Bouchkouj N, Kasamon YL, de Claro RA, George B, Lin X, Lee S, et al. FDA Approval Summary: Axicabtagene Ciloleucel for Relapsed or Refractory Large B-cell Lymphoma. Clinical Cancer Res. 2019;25:1702–8.
O’Leary MC, Lu X, Huang Y, Lin X, Mahmood I, Przepiorka D, et al. FDA Approval Summary: Tisagenlecleucel for Treatment of Patients with Relapsed or Refractory B-cell Precursor Acute Lymphoblastic Leukemia. Clin Cancer Res. 2019;25:1142–6.
Lu T, Ma R, Dong W, Teng K-Y, Kollath DS, Li Z, et al. Off-the-shelf CAR natural killer cells secreting IL-15 target spike in treating COVID-19. Nat Commun. 2022;13:2576.
Christodoulou I, Ho WJ, Marple A, Ravich JW, Tam A, Rahnama R, et al. Engineering CAR-NK cells to secrete IL-15 sustains their anti-AML functionality but is associated with systemic toxicities. J Immunother Cancer. 2021;9:e003894.
Daher M, Basar R, Gokdemir E, Baran N, Uprety N, Nunez Cortes AK, et al. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells. Blood. 2021;137:624–36.
Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38:947–53.
Zmievskaya E, Valiullina A, Ganeeva I, Petukhov A, Rizvanov A, Bulatov E. Application of CAR-T Cell Therapy beyond Oncology: Autoimmune Diseases and Viral Infections. Biomedicines. 2021;9:59.
Marofi F, Motavalli R, Safonov VA, Thangavelu L, Yumashev AV, Alexander M, et al. CAR T cells in solid tumors: challenges and opportunities. Stem Cell Res Ther. 2021;12:81.
Zhou T, Damsky W, Weizman O-E, McGeary MK, Hartmann KP, Rosen CE, et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature. 2020;583:609–14.
Aronovich EL, McIvor RS, Hackett PB. The Sleeping Beauty transposon system: a non-viral vector for gene therapy. Human Mol Genet. 2011;20:R14–R20.
Hurton LV, Singh H, Najjar AM, Switzer KC, Mi T, Maiti S et al. Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc Natl Acad Sci USA. 2016; 113. https://doi.org/10.1073/pnas.1610544113.
Tugues S, Burkhard SH, Ohs I, Vrohlings M, Nussbaum K, vom Berg J, et al. New insights into IL-12-mediated tumor suppression. Cell Death Differ. 2015;22:237–46.
Shum T, Omer B, Tashiro H, Kruse RL, Wagner DL, Parikh K, et al. Constitutive Signaling from an Engineered IL7 Receptor Promotes Durable Tumor Elimination by Tumor-Redirected T Cells. Cancer Discov. 2017;7:1238–47.
Huckaby JT, Landoni E, Jacobs TM, Savoldo B, Dotti G, Lai SK. Bispecific binder redirected lentiviral vector enables in vivo engineering of CAR-T cells. J Immunother Cancer. 2021;9:e002737.
Nawaz W, Huang B, Xu S, Li Y, Zhu L, Yiqiao H, et al. AAV-mediated in vivo CAR gene therapy for targeting human T-cell leukemia. Blood Cancer J. 2021;11:119.
Parayath NN, Stephan SB, Koehne AL, Nelson PS, Stephan MT. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat Commun. 2020;11:6080.
Smith TT, Stephan SB, Moffett HF, McKnight LE, Ji W, Reiman D, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotech. 2017;12:813–20.
Rurik JG, Tombácz I, Yadegari A, Méndez Fernández PO, Shewale SV, Li L, et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375:91–6.
Watanabe K, Nishikawa H. Engineering strategies for broad application of TCR-T- and CAR-T-cell therapies. Int Immunol. 2021;33:551–62.
Rath JA, Arber C. Engineering Strategies to Enhance TCR-Based Adoptive T Cell Therapy. Cells. 2020;9:1485.
Vazquez-Lombardi R, Jung JS, Schlatter FS, Mei A, Mantuano NR, Bieberich F, et al. High-throughput T cell receptor engineering by functional screening identifies candidates with enhanced potency and specificity. Immunity. 2022;55:1953–1966.e10.
Ng S, Tjhung KF, Paschal BM, Noren CJ, Derda R. Chemical Posttranslational Modification of Phage-Displayed Peptides. In: Derda R, editor. Peptide Libraries. Springer New York: New York, NY; 2015. p. 155–72. https://doi.org/10.1007/978-1-4939-2020-4_11.
Moesslacher CS, Kohlmayr JM, Stelzl U. Exploring absent protein function in yeast: assaying post translational modification and human genetic variation. Microb Cell. 2021;8:164–83.
Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503.
Wellner A, McMahon C, Gilman MSA, Clements JR, Clark S, Nguyen KM, et al. Rapid generation of potent antibodies by autonomous hypermutation in yeast. Nat Chem Biol. 2021;17:1057–64.
Funding
The authors acknowledge funding from National Institutes of Health (R01EY031097) and an Allegheny Health Network-Johns Hopkins Cancer Research Fund award. M.K. and K.P. are recipients of National Science Foundation Graduate Research Fellowship Program awards.
Author information
Authors and Affiliations
Contributions
M.K., K.P., L.M.T., and J.B.S. wrote and revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kizerwetter, M., Pietz, K., Tomasovic, L.M. et al. Empowering gene delivery with protein engineering platforms. Gene Ther 30, 775–782 (2023). https://doi.org/10.1038/s41434-022-00379-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00379-6