Abstract
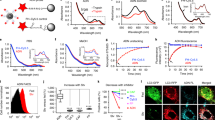
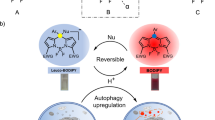
Autophagy is a cellular process with important functions that drive neurodegenerative diseases and cancers. Lysosomal hyperacidification is a hallmark of autophagy. Lysosomal pH is currently measured by fluorescent probes in cell culture, but existing methods do not allow for quantitative, transient or in vivo measurements. In the present study, we developed near-infrared optical nanosensors using organic color centers (covalent sp3 defects on carbon nanotubes) to measure autophagy-mediated endolysosomal hyperacidification in live cells and in vivo. The nanosensors localize to the lysosomes, where the emission band shifts in response to local pH, enabling spatial, dynamic and quantitative mapping of subtle changes in lysosomal pH. Using the sensor, we observed cellular and intratumoral hyperacidification on administration of mTORC1 and V-ATPase modulators, revealing that lysosomal acidification mirrors the dynamics of S6K dephosphorylation and LC3B lipidation while diverging from p62 degradation. This sensor enables the transient and in vivo monitoring of the autophagy–lysosomal pathway.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data are provided with this paper.
Code availability
LABVIEW code for data acquisition and MATLAB codes for data analysis in this article are available in the public GitHub repository (github.com/mijinee/HellerLab_MSKCC).
References
Lawrence, R. E. & Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019).
Mulcahy Levy, J. M. & Thorburn, A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 27, 843–857 (2020).
Galluzzi, L. et al. Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880 (2015).
Zhan, L. et al. Autophagy as an emerging therapy target for ovarian carcinoma. OncoTarget 7, 83476–83487 (2016).
Jung, S., Jeong, H. & Yu, S.-W. Autophagy as a decisive process for cell death. Exp. Mol. Med. 52, 921–930 (2020).
Lauzier, A. et al. Colorectal cancer cells respond differentially to autophagy inhibition in vivo. Sci. Rep. 9, 11316 (2019).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17, 1–382 (2021).
Bellot, G. et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581 (2009).
Chavez-Dominguez, R., Perez-Medina, M., Lopez-Gonzalez, J. S., Galicia-Velasco, M. & Aguilar-Cazares, D. The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity. Front. Oncol. 10, 578418 (2020).
Kimmelman, A. C. & White, E. Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043 (2017).
Janji, B., Berchem, G. & Chouaib, S. Targeting autophagy in the tumor microenvironment: new challenges and opportunities for regulating tumor immunity. Front. Immunol. 9, 887 (2018).
Mizushima, N. & Murphy, L. O. Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Sci. 45, 1080–1093 (2020).
Poillet-Perez, L. et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat. Cancer 1, 923–934 (2020).
Kuma, A., Komatsu, M. & Mizushima, N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 (2017).
Yoshii, SaoriR. et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev. Cell 39, 116–130 (2016).
Karsli-Uzunbas, G. et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 (2014).
Yim, W. W.-Y. & Mizushima, N. Lysosome biology in autophagy. Cell Discov. 6, 6 (2020).
Williams, R. M. et al. Harnessing nanotechnology to expand the toolbox of chemical biology. Nat. Chem. Biol. 17, 129–137 (2021).
Aref, M. et al. Potentiometric pH nanosensor for intracellular measurements: real-time and continuous assessment of local gradients. Anal. Chem. 93, 15744–15751 (2021).
Ma, L., Ouyang, Q., Werthmann, G. C., Thompson, H. M. & Morrow, E. M. Live-cell microscopy and fluorescence-based measurement of luminal pH in intracellular organelles. Front. Cell Dev. Biol. 5, 71 (2017).
Myochin, T. et al. Rational design of ratiometric near-infrared fluorescent pH probes with various pKa values, based on aminocyanine. J. Am. Chem. Soc. 133, 3401–3409 (2011).
Robinson, K. J. et al. Modified organosilica core–shell nanoparticles for stable pH sensing in biological solutions. ACS Sens. 3, 967–975 (2018).
Burgstaller, S. et al. pH-lemon, a fluorescent protein-based pH reporter for acidic compartments. ACS Sens. 4, 883–891 (2019).
Guha, S. et al. Approaches for detecting lysosomal alkalinization and impaired degradation in fresh and cultured RPE cells: evidence for a role in retinal degenerations. Exp. Eye Res. 126, 68–76 (2014).
Wäldchen, S., Lehmann, J., Klein, T., van de Linde, S. & Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 5, 15348 (2015).
Laissue, P. P., Alghamdi, R. A., Tomancak, P., Reynaud, E. G. & Shroff, H. Assessing phototoxicity in live fluorescence imaging. Nat. Methods 14, 657–661 (2017).
Heller, D. A. et al. Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proc. Natl Acad. Sci. USA 108, 8544–8549 (2011).
Kim, M. et al. Detection of ovarian cancer via the spectral fingerprinting of quantum-defect-modified carbon nanotubes in serum by machine learning. Nat. Biomed. Eng. 6, 267–275 (2022).
Bachilo, S. M. et al. Structure-assigned optical spectra of single-walled carbon nanotubes. Science 298, 2361 (2002).
Welsher, K., Sherlock, S. P. & Dai, H. Deep-tissue anatomical imaging of mice using carbon nanotube fluorophores in the second near-infrared window. Proc. Natl Acad. Sci. USA 108, 8943 (2011).
Mandal, A. K. et al. Fluorescent sp3 defect-tailored carbon nanotubes enable NIR-II single particle imaging in live brain slices at ultra-low excitation doses. Sci. Rep. 10, 5286 (2020).
Jena, P. V. et al. A carbon nanotube optical reporter maps endolysosomal lipid flux. ACS Nano 11, 10689–10703 (2017).
Galassi, T. V. et al. An optical nanoreporter of endolysosomal lipid accumulation reveals enduring effects of diet on hepatic macrophages in vivo. Sci. Transl. Med. 10, eaar2680 (2018).
Galassi, T. V. et al. Long-term in vivo biocompatibility of single-walled carbon nanotubes. PLoS ONE 15, e0226791 (2020).
Brozena, A. H., Kim, M., Powell, L. R. & Wang, Y. Controlling the optical properties of carbon nanotubes with organic colour-centre quantum defects. Nat. Rev. Chem. 3, 375–392 (2019).
Kwon, H. et al. Optical probing of local pH and temperature in complex fluids with covalently functionalized, semiconducting carbon nanotubes. J. Phys. Chem. C 119, 3733–3739 (2015).
Piao, Y. et al. Brightening of carbon nanotube photoluminescence through the incorporation of sp3 defects. Nat. Chem. 5, 840–845 (2013).
Gravely, M., Safaee, M. M. & Roxbury, D. Biomolecular functionalization of a nanomaterial to control stability and retention within live cells. Nano Lett. 19, 6203–6212 (2019).
Roxbury, D., Jena, P. V., Shamay, Y., Horoszko, C. P. & Heller, D. A. Cell membrane proteins modulate the carbon nanotube optical bandgap via surface charge accumulation. ACS Nano 10, 499–506 (2016).
Hornung, V. et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008).
Holtzmann, E. Lysosomes (Plenum, 1989).
Alberts, B. et al. Molecular Biology of the Cell, 4th edn (Garland Science, 2002).
Jin, H., Heller, D. A., Sharma, R. & Strano, M. S. Size-dependent cellular uptake and expulsion of single-walled carbon nanotubes: single particle tracking and a generic uptake model for nanoparticles. ACS Nano 3, 149–158 (2009).
Roxbury, D. et al. Hyperspectral microscopy of near-infrared fluorescence enables 17-chirality carbon nanotube imaging. Sci. Rep. 5, 14167 (2015).
Han, J. & Burgess, K. Fluorescent indicators for intracellular pH. Chem. Rev. 110, 2709–2728 (2010).
Yamamoto, A. et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 (1998).
Mauvezin, C. & Neufeld, T. P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome–lysosome fusion. Autophagy 11, 1437–1438 (2015).
Chung, C. Y.-S. et al. Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 15, 776–785 (2019).
Frost, L. S., Dhingra, A., Reyes-Reveles, J. & Boesze-Battaglia, K. The use of DQ-BSA to monitor the turnover of autophagy-associated cargo. Methods Enzymol. 587, 43–54 (2017).
Corrotte, M., Fernandes, M. C., Tam, C. & Andrews, N. W. Toxin pores endocytosed during plasma membrane repair traffic into the lumen of MVBs for degradation. Traffic 13, 483–494 (2012).
Peña-Llopis, S. et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 (2011).
Settembre, C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429 (2011).
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1*. J. Biol. Chem. 284, 8023–8032 (2009).
Sahani, M. H., Itakura, E. & Mizushima, N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 10, 431–441 (2014).
Harvey, J. D. et al. A carbon nanotube reporter of microRNA hybridization events in vivo. Nat. Biomed. Eng. 1, 0041 (2017).
Zhang, X.-X. et al. pH-sensitive fluorescent dyes: are they really pH-sensitive in cells? Mol. Pharm. 10, 1910–1917 (2013).
Subbaiyan, N. K. et al. Role of surfactants and salt in aqueous two-phase separation of carbon nanotubes toward simple chirality isolation. ACS Nano 8, 1619–1628 (2014).
Quintero, B., Cabeza, M. C., Martínez, M. I., Gutiérrez, P. & Martínez, P. J. Dediazoniation of p-hydroxy and p-nitrobenzenediazonium ions in an aqueous medium: interference by the chelating agent diethylenetriaminepentaacetic acid. Can. J. Chem. 81, 832–839 (2003).
Streit, J. K., Fagan, J. A. & Zheng, M. A low energy route to DNA-wrapped carbon nanotubes via replacement of bile salt surfactants. Anal. Chem. 89, 10496–10503 (2017).
Zheng, M. & Diner, B. A. Solution redox chemistry of carbon nanotubes. J. Am. Chem. Soc. 126, 15490–15494 (2004).
Bolte, S. & CordeliÈRes, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Acknowledgements
We thank Memorial Sloan Kettering Cancer Center (MSKCC) Molecular Cytology Core Facility for assistance with AFM imaging, sample processing for immunofluorescence and confocal microscopy, and image analysis. We also thank the Center for Translational Pathology at Weill Cornell Medicine for immunohistochemistry. We thank C. O’Mara at the Daniel Bachovchin lab (MSKCC) for the NLRP3 inflammasome experiment. We thank P. Jena for assistance with image processing and analysis. The graphical abstract, Fig. 3a and Supplementary Figs. 2 and 11 were created with BioRender.com. This work was supported in part by the National Science Foundation CAREER Award (grant no. 1752506 to D.A.H.), the National Cancer Institute (grant no. R01-CA215719 to D.A.H. and Cancer Center Support grant no. P30-CA008748 to D.A.H., H.A. and Y.M.L.), the National Institutes of Health (NIH) Common Fund (grant no. DP2-HD075698 to D.A.H.), the Department of Defense Congressionally Directed Medical Research Program (W81XWH2210563 to D.A.H.), the American Cancer Society Research Scholar grant (no. GC230452 to D.A.H.), the Ara Parseghian Medical Research Fund (to D.A.H.), the Honorable Tina Brozman Foundation (to D.A.H.), the Ovarian Cancer Research Alliance and the Edmée Firth Fund for Research in Ovarian Cancer (grant no. CRDGAI-2023-3-1003 to D.A.H.), the Pershing Square Sohn Cancer Research Alliance (to D.A.H.), the New York State Biodefense Commercialization Fund (to D.A.H.), the Expect Miracles Foundation—Financial Services Against Cancer (to D.A.H.), the Louis and Rachel Rudin Foundation (to D.A.H.), the Experimental Therapeutics Center of MSKCC (to D.A.H. and Y.M.L.), Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research (to D.A.H. and Y.M.L.), the JPB Foundation (to Y.M.L.) and the William Randolph Hearst Fund in Experimental Therapeutics (to Y.M.L.). M.K. was supported by the NIH (grant no. K99-EB033580) and the Marie-Josée Kravis Women in Science Endeavor Postdoctoral Fellowship. Z.Y. was supported by the Ann Schreiber Mentored Investigator Award (Ovarian Cancer Research Fund) and Young Investigator 2019 (Kaleidoscope of Hope). R.L. was supported by an NIH T32 training grant (no. T32-GM73546), J.W. was supported by an NIH T32 training grant (no. T32-GM136640-Tan). D.W. was supported by NIH T32 training grants (nos. T32-GM141949 and T32-CA062948). R.F. was supported by the Alfred Benzon Foundation Fellowship. Y.H.W. acknowledges support from the National Science Foundation (grant nos. CHE-1904488 and CHE-2204202).
Author information
Authors and Affiliations
Contributions
M.K., C. Chen, R.F., D.A.H., H.A. and Y.H.W. conceived the idea. M.K., C. Chen, R.F. and D.A.H. designed experiments. M.K., C. Chen, and D.A.H. analyzed the data. M.K., E.R. and X.W. performed nanosensor synthesis. M.K., C. Chen, R.F., E.R., J.W., C. Cupo, R.E.L., D.W.W., R.L. and D.G. performed in vitro experiments. M.K., C. Chen, Z.Y. and J.S. performed in vivo experiments. M.K., C. Chen and D.A.H. wrote the manuscript. Y.M.L., H.A. and Y.H.W. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.A.H. is a cofounder and officer with equity interest in Lime Therapeutics, Inc., cofounder with equity interest in Selectin Therapeutics Inc. and Resident Diagnostics, Inc., and a member of the scientific advisory board of Concarlo Therapeutics, Inc., Nanorobotics Inc. and Mediphage Bioceuticals, Inc. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Jiajie Diao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of OCC-DNA response in various buffer/media conditions.
Emission wavelengths of a-c, E11 and d-f, E11- of the OCC-DNA complexes at varying buffer pH and media conditions in phosphate buffered saline. The metal ion concentrations tested are physiologically relevant ranges. All data are presented as mean values and error bars denote standard deviation from N = 3 technical replicates (a-f). g, Frequency distribution of standard deviations of ΔE ( = E11- – E11) wavelength shifts of triplicate measurements of a-f.
Extended Data Fig. 2 Protein concentration effects on OCC-DNA optical response.
The dynamic range of the OCC-DNA response to pH at increasing concentrations of: a, bovine serum albumin and b, fetal bovine serum. All data are presented as mean values and error bars denote standard deviation from N = 3 technical replicates (a,b).
Extended Data Fig. 3 Viscosity effects on OCC-DNA optical response.
Emission wavelengths of a, E11 and b, E11- of the OCC-DNA complexes at varying buffer pH and glycerol. All data are presented as mean values and error bars denote standard deviation from N = 3 technical replicates (a,b).
Extended Data Fig. 4 Cell viability in response to OCC-DNA complexes.
Single-dose (0.01 mg/L) OCC-DNA cell viability of SKOV3, OVCAR3, HEK293, HeLa, MEF, RM1, and Myc-CaP cell lines, measured via CellTiter-Glo 2.0, after 72 hours of incubation. No statistically significant differences were observed between the PBS control groups (gray) and the treatment groups (red) in all the tested cell lines. All data are presented as mean values and error bars denote standard deviation of triplicates for each condition.
Extended Data Fig. 5 OCC-DNA responses in 8 cell lines.
The emission response (ΔE = E11- – E11) of OCC-DNAs in live cells upon exposure to HEPES or MES buffer solutions of varying pHs with monensin (see Methods). All data are presented as mean values and error bars denote standard deviation from N = 3–25 biological replicates.
Extended Data Fig. 6 Inhibitor-mediated alterations of nanosensor response to pH.
The emission response (ΔE = E11- – E11) of nanosensors in live SKOV3 cells upon exposure to HEPES or MES buffer solutions of varying pHs with monensin (see Methods). Cells were treated with DMSO (black), 100 µM EN6 (red), 100 nM bafilomycin A1 (blue), 250 nM torin 1 (green) for 4 hours prior to pH measurements. Data are presented as mean values and error bars denote standard deviation from N = 25 each DMSO, EN6, and Baf A1 point, and N = 25, 24, and 21 for pH 7, 5.06, and 3.16 for torin 1 as biological replicates.
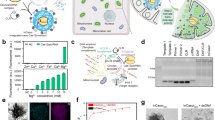
Extended Data Fig. 7 Nanosensor response in autophagy-defective cells.
a, ATG7 expression by western blotting confirmed the knockout of ATG7 in the HEK293T cell line used herein. The emission wavelength response (ΔE = E11- – E11) of OCC-DNAs in live b, wild type and c, ATG7-/- HEK293T cells upon exposure to HEPES or MES buffer solutions of varying pHs in the presence of monensin (see Methods). Cells were treated with DMSO (black) or 250 nM torin 1 (blue) for 4 hours prior to pH measurements. All data are presented as mean values and error bars denote standard deviation from N = 10 technical replicates (b,c). Original gel images are in Supplementary Fig. 24.
Extended Data Fig. 8 Time-dependent fluorescence intensity changes of intratumorally-injected nanosensors.
Quantification of total emission intensity of nanosensors in solid tumours of mice after injection. Fluorescence measurements were performed with a near-infrared preclinical hyperspectral imager with 730 nm excitation. Data are presented as mean values and error bars denote standard deviation from N = 5 biological replicates.
Supplementary information
Supplementary Information
Supplementary Figs. 1–24 and Supplementary References.
Supplementary Video 1
NIR fluorescence video of OCC–DNA complexes in live SKOV3 cells.
Supplementary Data 1
Source data for Supplementary Figs.
Source data
Fig. 1
Statistical source data.
Fig. 2
Statistical Source Data
Fig. 3
Statistical source data.
Fig. 4
Statistical source data.
Fig. 4
Unprocessed western blots/gels for actin, LC3B, P62 and pS6K.
Fig. 5
Statistical source data.
Fig. 6
Statistical source data.
Extended Data Fig. 1
Statistical source data.
Extended Data Fig. 2
Statistical source data.
Extended Data Fig. 3
Statistical source data.
Extended Data Fig. 4
Statistical source data.
Extended Data Fig. 5
Statistical source data.
Extended Data Fig. 6
Statistical source data.
Extended Data Fig. 7
Statistical source data.
Extended Data Fig. 7a
Unprocessed western blots/gels for actin and FIP200.
Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, M., Chen, C., Yaari, Z. et al. Nanosensor-based monitoring of autophagy-associated lysosomal acidification in vivo. Nat Chem Biol 19, 1448–1457 (2023). https://doi.org/10.1038/s41589-023-01364-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-023-01364-9
This article is cited by
-
Near-infrared II fluorescence imaging
Nature Reviews Methods Primers (2024)
-
Ratiometric fluorescent sensing of pyrophosphate with sp³-functionalized single-walled carbon nanotubes
Nature Communications (2024)
-
Lysosomal acidification dysfunction in microglia: an emerging pathogenic mechanism of neuroinflammation and neurodegeneration
Journal of Neuroinflammation (2023)
-
Human and environmental safety of carbon nanotubes across their life cycle
Nature Reviews Materials (2023)
-
Monitoring lysosomal acidity
Nature Chemical Biology (2023)