Abstract
The E3 ligase TRIM7 has emerged as a critical player in viral infection and pathogenesis. However, the mechanism governing the TRIM7–substrate association remains to be defined. Here we report the crystal structures of TRIM7 in complex with 2C peptides of human enterovirus. Structure-guided studies reveal the C-terminal glutamine residue of 2C as the primary determinant for TRIM7 binding. Leveraged by this finding, we identify norovirus and SARS-CoV-2 proteins, and physiological proteins, as new TRIM7 substrates. Crystal structures of TRIM7 in complex with multiple peptides derived from SARS-CoV-2 proteins display the same glutamine-end recognition mode. Furthermore, TRIM7 could trigger the ubiquitination and degradation of these substrates, possibly representing a new Gln/C-degron pathway. Together, these findings unveil a common recognition mode by TRIM7, providing the foundation for further mechanistic characterization of antiviral and cellular functions of TRIM7.

Similar content being viewed by others
Main
The tripartite motif (TRIM) family is a rapidly expanding protein superfamily with more than 80 genes identified in human1,2,3,4,5. The TRIM family proteins play essential roles in a plethora of cellular processes, including signal transduction, oncogenesis, cell death and antimicrobial responses1,3,4,5,6,7. While most TRIM proteins are characterized by the conserved RBCC domain architecture consisting of the RING (R), one or two B-boxes (B) and the coiled-coil (CC) domains, and a variable C-terminal domain (CTD)2,4,5, eight non-canonical members have been found lacking the RING-finger domain1. In the RBCC architecture, the RING domain recruits ubiquitin-loaded E2 and catalyzes the conjugation of ubiquitin to substrates, and the B-box and coiled-coil domains mainly play regulatory roles in the oligomerization and ligase activities. The CTD usually confers substrate specificity. More than ten distinct types of CTD in TRIM proteins have been identified, including the PRY-SPRY domain, which is present only in vertebrates7,8. The PRY-SPRY domain (also known as B30.2), a protein–protein interaction module widely present in eukaryotic proteins, features a twisted β-sandwich fold composed of two β-sheets8. About half of TRIM proteins possess a PRY-SPRY domain at the C terminus2, which may recruit substrates for ubiquitination by the RING domain.
TRIM7, featured with one B-box domain in the RBCC architecture and a C-terminal PRY-SPRY domain, is implicated in multiple critical cellular processes, such as glycogen biosynthesis and oncogenic signaling pathways, including c-Jun/AP1 activation and the DUSP6/p38 pathway9,10,11. Akin to other members in the TRIM family such as TRIM21, TRIM25 and TRIM56 (refs. 6,7), TRIM7 has been reported to mediate different types of ubiquitination modifications that dictate the differential fates of substrates. For example, while overexpression of TRIM7 causes K48-linked ubiquitination and degradation of DUSP6 (ref. 11), TRIM7 induces K63-linked ubiquitination and stabilization of RACO-1 protein in the c-Jun/AP1 pathway10. Notably, TRIM7 is also emerging as a crucial player in viral pathogenesis12,13,14,15,16. A recent study has identified human TRIM7 as an intrinsic antiviral effector against human enteroviruses such as coxsackievirus B3 (CVB3)12. TRIM7-mediated ubiquitination and subsequent degradation of CVB3 2BC protein are implicated to inhibit viral infection. The non-structural protein 2C of enteroviruses, which is derived from proteolysis of the precursor 2BC protein, functions as an ATPase/helicase to regulate viral RNA replication. TRIM7 recognizes the C-terminal region of 2C via PRY-SPRY domain, leading to ubiquitination and proteasomal degradation of viral 2BC protein but not 2C protein, thereby suppressing enterovirus replication12. In addition, a genome-wide CRISPR screen reveals that TRIM7 restricts norovirus infection in a ubiquitination-dependent manner through as-yet unknown substrates14.
To elucidate the molecular mechanism of substrate recognition by TRIM7, we have determined multiple crystal structures of TRIM7 in complex with peptides derived from CVB3 2C. The structural studies combined with biochemical characterization allow us to propose a glutamine-end motif binding mechanism, where TRIM7 recognizes 2C mainly through C-terminal glutamine-specific interactions. Strikingly, this binding mode is found to be preserved for other targets. More importantly, leveraged by this new knowledge, we are able to predict and identify new viral targets in noroviruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus as well as new physiological targets. Our findings not only unveil a general substrate-binding principle for TRIM7 but also suggest a possible role of TRIM7 in the degradation of viral and cellular proteins, potentially opening new avenues for therapeutic intervention in viral diseases.
Results
Biochemical characterization of TRIM7 and 2C association
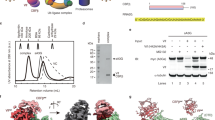
Human TRIM7 restricts CVB3 enterovirus infection by recognizing and degrading the viral protein 2BC, which could be further proteolytically processed to 2B and 2C proteins12. The physical association was indicated to primarily occur between the PRY-SPRY domain of TRIM7 (TRIM7PRY-SPRY, amino acids 338–511) and the 2C moiety of 2BC, especially the C-terminal region of 2C12 (Fig. 1a). It is challenging to purify the full-length TRIM7 and 2C proteins owing to solubility. To further characterize the TRIM7–2C interaction, we therefore purified TRIM7PRY-SPRY and the N-terminal-truncated 2C (2CΔN, amino acids 117–329) and then examined their binding using isothermal titration calorimetry (ITC). Our ITC results showed that TRIM7PRY-SPRY binds to 2CΔN with a dissociation constant (KD) of ~6 μM (Fig. 1b). By contrast, no obvious binding was observed after truncating the last 11 residues of 2C (amino acids 117–318), consistent with the previous pull-down assay results12. In line with these results, the C-terminal-derived peptide fragment displayed a KD value of ~3 μM toward TRIM7PRY-SPRY (Fig. 1b), comparable to that of 2CΔN protein, demonstrating a dominant role of this C-terminal motif in TRIM7 binding.
a, Domain organization of TRIM7 and 2C. NTD, N-terminal domain; ZnF, zinc finger. b, ITC measurements of 2C into TRIM7PRY-SPRY. ND, no detectable binding. c, Crystal structure of TRIM7PRY-SPRY–2C peptide complex. The bound 2C peptide is colored in green. The TRIM7 PRY-SPRY domain is composed of two antiparallel β-sheets (β-sheet A in orange, β-sheet B in violet red), connected with loops. Positions of loop 3 and loop 6 are indicated by arrows. d, The electrostatic potential of the TRIM7 PRY-SPRY domain.
Apart from the type B enterovirus CVB3, TRIM7 was also reported to inhibit other human enteroviruses such as enterovirus 71 (type A), poliovirus (type C) and enterovirus D68 (type D)12. Sequence analysis of these 2C proteins revealed a highly conserved C terminus (Extended Data Fig. 1a), indicating that TRIM7 may use a general binding mode for different 2C proteins of human enteroviruses. This is supported by the results that TRIM7 binds to the C-terminal peptides of different 2C proteins with similar affinities (KD values in the range 3–9 μM) (Extended Data Fig. 1b).
Overall structure of the TRIM7PRY-SPRY–2C peptide complex
Next, we set out to further elucidate the structural basis of TRIM7–2C association. Although extensive crystallization trials of TRIM7PRY-SPRY–2CΔN complex failed, we were able to crystallize and determine multiple structures of TRIM7PRY-SPRY in complex with peptides derived from CVB3 2C (amino acids 319-329) at high resolutions (1.1–1.6 Å) (Supplementary Table 1). Atomic protein models were built unambiguously for TRIM7PRY-SPRY and the last four to ten residues of 2C peptide (Fig. 1c and Extended Data Fig. 2a). All the solved structures with different space groups are nearly identical (Extended Data Fig. 2b), excluding the possibility of artifacts caused by crystal packing. TRIM7PRY-SPRY exhibits a typical PRY-SPRY domain fold8, composed of two antiparallel β-sheets connected by loops of variable length (Fig. 1c). These two β-sheets, β-sheet A and β-sheet B, pack against each other in a twisted β-sandwich configuration with a concave surface on one side and a convex surface on the other side. The β-sheet A and loops 3 and 6 form a positively charged groove to accommodate 2C (Fig. 1d). In comparison to the structure of TRIM7PRY-SPRY alone, there is no obvious conformational change upon 2C binding17 (Extended Data Fig. 2b). The binding affinity between TRIM7PRY-SPRY and 2CΔN was reduced in a higher-salt binding buffer, demonstrating that the polar contacts contribute to their physical association (Extended Data Fig. 2c). Mapping of the evolutionary sequence conservation to TRIM7PRY-SPRY structure revealed an invariant binding pocket18 (Extended Data Fig. 2d), suggesting a conserved 2C recognition mode across TRIM7 orthologs.
Interaction of TRIM7 with 2C
In the present structure, the C terminus of 2C displays a helical structure that inserts into the binding groove on the concave side of TRIM7PRY-SPRY. Specifically, the C-terminal Gln329 (termed as position −1) of 2C situated at the bottom of the binding pocket is involved in substantial interactions with TRIM7 (Fig. 2a). The side chains of Asn383, Arg385 and Ser499 from TRIM7 make polar interactions with the backbone carboxyl group of Gln329. On the other side, the side chain amide moiety of Gln329 is buttressed by hydrogen bonds to the main chains of Gly408 and Ser499, as well as the side chains of Gln436 and Ser499 from TRIM7. The side chain of Gln329 is further bracketed by Phe426 and Cys501 through hydrophobic interactions.
a, The C-terminal Q329 inserts into a pocket formed by residues N383, R385, G408, F426, Q436, S499 and C501 in TRIM7. b, The interaction interface between the two residues preceding Q329 in 2C (F328 and L327) and TRIM7. c, The interface between 2C residues 320–326 and TRIM7. d,e, ITC measurements of TRIM7PRY-SPRY mutants with 2C. f, ITC measurements of TRIM7PRY-SPRY mutants with 2C mutants.
The backbone carbonyl group of Phe328 (position −2) of 2C engages in hydrogen bonds with Thr384 of TRIM7 (Fig. 2b), while its aromatic ring is further stabilized by hydrophobic contacts with Thr382 and Leu423. Leu327 (position −3) of 2C is anchored in place mainly through the hydrophobic interaction with Leu423 of TRIM7. Nevertheless, few contacts were observed beyond the last three residues of 2C (Fig. 2c).
Mutagenesis studies of the TRIM7–2C binding interface
To explore the importance of the observed interactions, we introduced point mutations into TRIM7PRY-SPRY and measured their binding affinities. Replacement of the Gln329-interacting residues with alanine, including Asn383, Arg385, Phe426, Gln436, Ser499 and Cys501, greatly reduced or abolished 2CΔN binding (Fig. 2d and Supplementary Fig. 1). Similarly, the TRIM7PRY-SPRY–2CΔN interaction was disrupted when mutating the Phe328-interacting residues (Thr382, Thr384 and Leu423) (Fig. 2e and Supplementary Fig. 1). However, the substitution of other residues (such as Asn438 and Ser502) in the binding groove with alanine had little or no effect on the binding affinity (Supplementary Fig. 1).
To further discern the residues of 2C crucial for TRIM7 binding, we generated point mutations on 2CΔN for binding studies. Not surprisingly, both the Q329A and F328A mutations greatly impaired the binding (Fig. 2f and Supplementary Fig. 2). Nonetheless, alanine substitutions of residues Leu327–Thr322 had no obvious impact on binding. These results further confirmed the critical role of the last two residues of 2C, Gln329 and Phe328, in TRIM7 binding. As previously reported, alanine mutation of Thr323 at 2C would confer resistance to TRIM7 (ref. 12). In our structure, the hydroxyl group of Thr323 is hydrogen bonded to Asn438 of TRIM7 (Fig. 2c). However, mutations of Thr323, including T323A, T323G, T323S and T323I, had little effect on the binding (Extended Data Fig. 3 and Supplementary Fig. 2). One proposed possibility is that these variants would alter the plasticity and enzymatic activity of 2C to gain a fitness advantage in vivo12.
TRIM7 recognizes the C-terminal glutamine-containing motif
To further understand the binding preference of TRIM7, we generated a series of 2C mutants on the last three positions and examined their binding. Gln329 at position −1 fits snugly into the binding pocket (Fig. 3a). Substitution of Gln329 with bulkier residues, such as tyrosine, arginine and methionine, which is expected to have steric clashes, exhibited no binding to TRIM7PRY-SPRY (Supplementary Fig. 3a). Likewise, replacement of Gln329 with smaller residues (alanine, threonine, asparagine and valine) disrupted TRIM7PRY-SPRY binding, which is likely due to loss of the interactions mediated by side chain amide groups. Notably, mutation of Gln329 to glutamate with a highly similar chemical structure also abrogated the binding. In agreement with the ITC measurement, molecular dynamics (MD) simulation analysis of the complex structure revealed a strong tendency of Q329E mutant peptide to dissociate from TRIM7PRY-SPRY (Extended Data Fig. 4a, b). This may be due to that the negatively charged side chain carboxyl group of glutamate is more prone to hydrogen bonding with water molecules, which disrupts the interaction with TRIM7PRY-SPRY (Supplementary Table 2). The free C-terminal backbone carboxyl group is also critical for TRIM7PRY-SPRY binding, which is supported by the finding that no detectable binding was observed in the presence of the C-terminal amidation modification (Supplementary Fig. 3a). Collectively, these results suggest that a glutamine residue at position −1 is indispensable and irreplaceable for TRIM7 binding.
a, The C-terminal glutamine-binding pocket in TRIM7. The key residues located in the pocket are shown as stick representations (colored orange). Q329 (C terminus, −1 position) of 2C is colored in green. b, The pocket in TRIM7 accommodating F328 (−2 position) and L327 (−3 position) of 2C (colored green). c, Integrated heat plots for ITC measurements of TRIM7PRY-SPRY with peptide fragments of 2C protein in non-human enterovirus MenV.
Phe328 at position −2 rests against the rim of the pocket with its aromatic ring surrounded by the hydrophobic side chains of Leu423, Thr382 and Thr384 (Fig. 3b). Replacement of Phe328 with smaller residues (alanine, threonine, valine, asparagine and glutamate), with the potential to lose the phenyl-group-mediated hydrophobic interaction, substantially reduced TRIM7PRY-SPRY binding (Supplementary Fig. 3b,c). Arginine substitution of Phe328 would also weaken the interaction owing to the electrostatic repulsion with Arg354 of TRIM7, which agrees with our finding that F328R mutation greatly diminished the binding of TRIM7PRY-SPRY. However, substitution of Phe328 with bulky residues such as methionine and tyrosine would be better tolerated. Indeed, F328M had a similar binding affinity to TRIM7PRY-SPRY as wild type (WT) (Supplementary Fig. 3b,c). Leu327 at position −3 protrudes outwards from the pocket (Fig. 3b), forming loose interactions with the binding surface of TRIM7. Different types of amino acid substitutions at position −3 were all tolerated (Supplementary Fig. 4). Taken together, our results suggest that TRIM7 not only specifically recognizes glutamine at position −1, but also has a preference for bulky hydrophobic residues at position −2. This observation could explain why 2C protein from non-human enterovirus mengovirus (MenV), which harbors an alanine residue at the position equivalent to Phe328 in CVB3 2C protein, fails to interact with TRIM7 (ref. 12). Mutating alanine to phenylalanine in 2C of MenV was sufficient to restore the TRIM7 binding (Fig. 3c), highlighting the importance of the secondary determinant involving −2 position.
The potential norovirus substrate for TRIM7
TRIM7 was recently reported to inhibit the replication of norovirus14. However, the target of TRIM7 remains poorly understood. Referring to the observed glutamine-end motif binding rule, we therefore examined the sequence information of norovirus proteins. Several viral proteins, such as capsid protein VP1, p48, NTPase and the 3C-like protease, were found bearing the TRIM7-recognition motif, among which NTPase was found most conserved in different genogroups (GI–GV) (Fig. 4a). We therefore focused our study on NTPase.
a, Multiple sequence alignment of norovirus NTPase proteins from different genogroups (GI–GV). The aligned C-terminal last 10 residues are shown. The GI–GIV NTPase proteins are derived from human noroviruses and the GV NTPase protein originates from murine norovirus. Invariant and conserved residues are shaded green and yellow, respectively. b, Thermodynamic analysis of the interaction between TRIM7PRY-SPRY and NTPase peptide fragments from GI (left) and GV (middle), and GV 3C-like protease (right). c, Human norovirus NTPase co-immunoprecipitated with TRIM7. FLAG-NTPase or empty vector was co-transfected with HA-TRIM7 in HEK293T cells. HA-TRIM7-Mut, TRIM7 catalytic dead mutant (C29A/C31A). d, Expression of TRIM7-induced ubiquitination of human norovirus NTPase. FLAG-NTPase or empty vector was co-transfected with Ubi-Myc in the presence or absence of HA-TRIM7 in HEK293T cells. e, TRIM7 promoted the degradation of NTPase in a dose-dependent fashion. Mut, TRIM7 catalytic dead mutant. f, IFNB1 promoter reporter assay. HEK293T cells were co-transfected with an IFNB1-promoter-driven luciferase reporter construct, a RIG-I CARD expression plasmid and the indicated NTPase expression plasmid in the presence or absence of TRIM7. Relative luciferase activity was quantified 24 h after transfection. Data are presented as mean ± s.e.m.; n = 8, biologically independent samples were examined over three independent experiments. Statistical significance was determined by comparing each respective ‘RIG-I CARD + Vector’ group using two-way ANOVA with Tukey’s correction. ****P < 0.0001. n.s., not significant.
ITC measurement showed that the NTPase C-terminal peptide fragments from both human (GI) and murine (GV) noroviruses directly interact with TRIM7PRY-SPRY with KD values of ~6–10 µM (Fig. 4b), while alanine replacement of the C-terminal glutamine abolished the binding (Supplementary Fig. 5). Consistently, WT but not the C-terminal alanine-substituted NTPase protein (NTPase-Q/A), co-immunoprecipitated with TRIM7 when co-expressed in HEK293T cells (Fig. 4c). These results suggest that TRIM7 indeed can recognize norovirus NTPase using the glutamine-end motif binding mechanism both in vitro and in cells.
We next asked whether TRIM7 could ubiquitinate and degrade norovirus NTPase. As expected, WT TRIM7 but not the ligase dead mutant (TRIM7-mut), robustly ubiquitinated NTPase (Fig. 4d). Furthermore, expression of WT TRIM7 triggered NTPase degradation in a dose-dependent manner, whereas the degradation was inhibited by treatment with the proteasome inhibitor MG132 (Fig. 4e), implicating the 26S proteasome-dependent degradation. It has been demonstrated that norovirus NTPase impairs interferon-β (IFN-β) expression19. We next determined whether TRIM7 interferes with this NTPase-mediated suppression of the immune response. The expression of NTPase inhibited the IFNB1 promoter activity in the luciferase assay, whereas WT TRIM7 restored this activity (Fig. 4f and Supplementary Fig. 6a,b). However, the ligase dead TRIM7 mutant had no obvious effect on IFNB1 induction. The NTPase-mediated suppression of IFNB1 promoter activity was not affected regardless of the presence or absence of WT TRIM7 when the C-terminal glutamine of NTPase was mutated to alanine (Fig. 4f), further indicating the importance of C-terminal glutamine-specific recognition. We further analyzed the secreted IFN-β protein by enzyme-linked immunosorbent assay (ELISA). Consistently, the IFN-β protein level inhibited by NTPase was greatly increased in the presence of TRIM7 (Supplementary Fig. 7).
TRIM7 recognizes and ubiquitinates SARS-CoV-2 proteins
As TRIM7 restricts the infection of human enterovirus and norovirus12,14, both of which are positive single-stranded RNA (+ssRNA) viruses, it raises an interesting question whether TRIM7 could function as a broad-spectrum regulator in +ssRNA virus infection. We therefore attempted to explore whether another +ssRNA virus, SARS-CoV-2, could be targeted by TRIM7. Sequence analysis was performed for proteins encoded by SARS-CoV-2 genome, surprisingly revealing that most of the non-structural proteins (NSPs), including NSP4–10, NSP12–15, and the structural membrane (M) protein, have the glutamine-end motif (Fig. 5a). ITC experiments were then performed to evaluate their association with TRIM7PRY-SPRY. As revealed, binding was detected for all the measured peptides derived from the C termini of these candidates (Supplementary Fig. 8a,b). To further confirm the interaction, full-length NSP5 protein was purified, which consistently was found bound with TRIM7PRY-SPRY (Supplementary Fig. 8c). These were in agreement with the co-immunoprecipitation experiment, where WT NSP5 and NSP8, but not the mutants with alanine substitutions at the C-terminal glutamine (Q/A), were detected in the immunoprecipitated samples of TRIM7 (Extended Data Fig. 5a). Furthermore, expression of TRIM7 triggered the ubiquitination and proteasomal degradation of NSP5 and NSP8, whereas the degradation was inhibited when the gene encoding TRIM7 was knocked out (Fig. 5b–d and Supplementary Fig. 9). We further determined the crystal structures of TRIM7PRY-SPRY in complex with NSP5-, NSP8- and NSP12-derived peptides at high resolutions (1.2–1.4 Å) (Supplementary Table 3) (Fig. 5e,f and Extended Data Fig. 5b,c). Although the C-terminal sequences among these three NSP proteins are divergent (Fig. 5a), their binding modes to TRIM7 are similar to that of 2C, confirming the conserved C-terminal glutamine recognition mechanism by TRIM7. Together, these results suggest that TRIM7 could potentially target a large array of SARS-CoV-2 viral proteins, such as NSP5 and NSP8, for ubiquitination and proteasomal degradation via the glutamine-end motif binding mechanism.
a, Multiple sequence alignment of SARS-CoV-2 viral proteins ended with glutamine at the C terminus. Invariant and conserved residues are shaded green and yellow, respectively. b, In cell ubiquitination assay for NSP5 and NSP8. FLAG-tagged NSP5, NSP8 or empty vector were co-transfected with Ubi-Myc in the presence or absence of HA-TRIM7 in HEK293T cells. HA-TRIM7-Mut, TRIM7 catalytic dead mutant (C29A/C31A). c,d, TRIM7 induced the proteasomal degradation of NSP5 (c) and NSP8 (d) in a dose-dependent fashion. Cellular degradation assays in cells expressing the HA-TRIM7 and FLAG-NSP5/8 proteins as indicated. e,f, Overall structures of TRIM7PRY-SPRY in complex with NSP5 (amino acids 300–306; e) or NSP8 (amino acids 194–198; f). g, IFNB1 promoter reporter assay. HEK293T cells were co-transfected with an IFNB1-promoter-driven luciferase reporter construct, a RIG-I CARD plasmid and the indicated NSP protein expression plasmids with or without the TRIM7 expression plasmid. Relative luciferase activity was quantified 24 h after transfection. Data are presented as mean ± s.e.m.; n = 6, biologically independent samples were examined over three independent experiments. Statistical significance was determined by comparing each respective ‘RIG-I CARD + Vector’ group using two-way ANOVA with Tukey’s correction. ****P < 0.0001.
SARS-CoV-2 proteins were reported to antagonize IFN signaling20,21,22,23,24,25. Consistently, we observed that expression of SARS-CoV-2 viral proteins, including NSP5, NSP6 and NSP8, greatly suppressed IFNB1 promoter activity and IFN-β production (Extended Data Fig. 5d and Supplementary Fig. 6c). However, IFNB1 promoter activity was rescued in the presence of WT TRIM7 (Fig. 5g and Supplementary Figs. 6d and 7). Similarly, expression of TRIM7 also restored TBK1 phosphorylation, a key event for IFN-β signaling that was impaired by NSP5 or NSP8 protein (Extended Data Fig. 5e,f). To reconcile the glutamine-end binding mode, we also introduced mutation into the C-terminal glutamine residues in NSP5 and NSP8 (Q/A). These mutants blocked the IFNB1 promoter induction and TBK1 phosphorylation similar to the WT NSP5 and NSP8 (Fig. 5g and Extended Data Fig. 5e,f). Nonetheless, TRIM7 failed to restore IFNB1 promoter activity in the presence of these NSP mutations. Together, our data suggest that TRIM7 may function to promote the degradation of viral proteins of norovirus and SARS-CoV-2, thereby restoring the type-I interferon response inhibited by these proteins.
Recognition of cellular proteins by TRIM7
We next asked whether the same C-terminal glutamine motif recognition mechanism used by TRIM7 is adopted by the cellular targets as well. Multiple cellular proteins that have been previously reported as putative targets of TRIM7 were explored, including BRMS1, DUSP6, GN1, RACO-1 and STING10,11,15,26,27. Indeed, TRIM7PRY-SPRY was able to directly interact with the C-terminal peptide fragments of GN1 (~ 8 µM) and RACO-1 (~ 10 µM), both of which contain tolerable glutamine-end motifs (Extended Data Fig. 6a). By contrast, the peptides or protein fragments, which are derived from the C termini of BRMS1, DUSP6 and STING without glutamine ends, exhibited no obvious binding toward TRIM7PRY-SPRY (Extended Data Fig. 6b). The potential explanation is that these proteins could possibly interact with TRIM7 either through a different interface or additional binding partners16,28.
In addition, RWDD2B protein featuring a C-terminal glutamine is suggested as the putative interaction partner of TRIM7 by mining the available protein–protein interaction databases29. ITC measurements confirmed its direct binding with TRIM7PRY-SPRY, indicative of a new potential substrate (Fig. 6a). Consistently, further cellular experiments for RWDD2B confirmed that it could be co-immunoprecipitated with TRIM7 in a C-terminal glutamine-dependent fashion (Fig. 6b). TRIM7 overexpression induced efficient ubiquitination of RWDD2B (Fig. 6c and Supplementary Fig. 10). The ubiquitination of RWDD2B was blocked when expressing the K48R ubiquitin mutant but not K11R or K63R, implying that TRIM7 may primarily catalyze the formation of K48-linked ubiquitin chains on RWDD2B (Supplementary Fig. 11). As anticipated, we observed the degradation of RWDD2B in the presence of TRIM7 (Fig. 6d and Supplementary Fig. 9). Additionally, the RWDD2B protein level decreased in HEK293T cells in the cycloheximide chase assays in the presence of TRIM7 (Fig. 6e). Nevertheless, both overexpressed and endogenous RWDD2B protein remained stable in the TRIM7-knockout cells (Fig. 6e and Supplementary Fig. 9). Collectively, these data support the notion that RWDD2B degradation is mediated by TRIM7 via the glutamine-end specific binding.
a, ITC measurement of TRIM7PRY-SPRY with peptide fragments derived from the C terminus of RWDD2B. b, Co-immunoprecipitation of RWDD2B with TRIM7. RWDD2B-Q/A, the alanine replacement of the C-terminal glutamine in RWDD2B. c, In cell ubiquitination assay for RWDD2B. d, TRIM7 induced proteasomal degradation of RWDD2B. Cellular degradation assays in cells expressing the HA-TRIM7 and FLAG-RWDD2B proteins as indicated. e, Top, cycloheximide chase analysis of RWDD2B in TRIM7-knockout (TRIM7−/−), WT and TRIM7 overexpression (TRIM7ov) HEK293T cells. Bottom, the ratios of RWDD2B to β-actin are plotted.
Discussion
Our data reveal the C-terminal glutamine-end as the primary determinant for TRIM7 binding. TRIM7 also displays a strong preference for bulky hydrophobic amino acids at position −2. Leu423 in TRIM7, which is embraced by the two residues at −2 and −3 positions in substrate (Figs. 2b and 3b), is essential for substrate binding; replacement with alanine or valine, with the potential to weaken hydrophobic contacts, abolished substrate binding (Supplementary Fig. 1), implying that Leu423 may work as a hydrophobic sensor to fine tune the substrate binding and preference. For example, obvious binding was observed for RWDD2B (Fig. 6a), which bears a small glycine residue at position −2. Notably, RWDD2B contains an unbranched long side chain residue at position −3 (glutamate) (Fig. 6a). It is plausible that the bulky residue at position −3 would provide a compensation for the loss of bulky residue at position −2. Other positions of substrate may also act synergistically to affect the TRIM7 binding.
We identified the norovirus NTPase, which harbors a highly conserved glutamine-end across genogroups, as a new substrate of TRIM7. Ubiquitination and proteasomal degradation of norovirus NTPase could potentially suppress the enzymatic activities necessary for viral replication30, and counteract its negative effect on IFN-β production (Fig. 4f and Supplementary Fig. 7)19, which may, at least partially, explain the antiviral activity of TRIM7 against norovirus14. In addition, the 3C-like protease possessing a C-terminal glutamine residue in murine norovirus but not in other noroviruses could possibly be an extra, murine norovirus-specific TRIM7 target (Fig. 4b). Through the same glutamine-end motif recognition mechanism, we found TRIM7 might also be able to target a large fraction of SARS-CoV-2 proteins, including the structural protein M, non-structural proteins NSP4–10 and NSP12–15, all of which are essential proteins for SARS-CoV-2 viral replication31,32. Moreover, most of the glutamine-end motifs in the full-length protein structures are likely to be accessible to TRIM7, such as NSP5 and the NSP8-containing replication–transcription complex32,33. Further virology studies will be needed to explore the potential role of TRIM7 in SARS-CoV-2 infection. TRIM7 was also reported to inhibit IFN-β production in response to vesicular stomatitis virus (VSV) infection in THP1 and BMDM cells16 and found to be involved in facilitating Zika virus (+ssRNA) cell entry by promoting the ubiquitination of the viral envelope protein13, suggesting multifaceted roles of TRIM7 in viral infection. Collectively, these findings imply potentially prevalent and versatile roles of TRIM7 E3 ligase in +ssRNA virus infection.
The +ssRNA viral genomes generally encode polyproteins, which are proteolytically cleaved into individual structural and non-structural viral proteins by viral proteases. The substrate cleavage specificity of these viral proteases therefore determines the C-terminal sequences of the mature viral proteins. The 3C or 3C-like proteases of human enterovirus, norovirus and SARS-CoV-2 virus show cleavage preference for the glutamine at the position P1 (refs. 34,35,36), releasing glutamine-terminated viral proteins, which explains the vulnerability of their mature viral proteins to TRIM7. Therefore, examination of more viral proteases may further extend the spectrum of +ssRNA viruses targeted by TRIM7.
Interestingly, on the basis of the glutamine-end motif recognition mechanism, we further predicted and identified RWDD2B as a new cellular substrate of TRIM7. RWDD2B is suggested as a target of osteoarthritis susceptibility37, implying more diverse functional roles of TRIM7. As there are ~1,000 human proteins terminated with glutamine, the recognition principle described here would potentially enable the identification of more physiological targets of TRIM7.
The ubiquitin–proteasome system controls protein homeostasis through recognizing degradation signals (degrons) identified in the N or C terminus of proteins, namely N- or C-degrons, respectively38. Several classes of C-degrons such as Arg/C-end, Glu/C-end, Gly/C-end and Ala/C-end have recently been discovered39,40. Analogous to the C-degrons recognition by the receptor in E3 ligases41,42,43,44,45, the C terminus of the glutamine-end motifs embeds deeply in a positively charged pocket of TRIM7. Interestingly, TRIM7 appears to recognize targets with a helical conformation (Figs. 1c and 5e,f, and Extended Data Fig. 2a), akin to FBXO31 targeting cyclin D1 C-degron44. The peptides derived from 2C, NSP5 and NSP12 but not NSP8 show helix preference in isolation, while all these peptides display helical conformation in our structures. Thus, it is likely that helical configuration could be induced upon TRIM7 binding. Although not all the targets undergo the degradative K48-linked polyubiquitination9,10,12, the observed C-end glutamine rule by the TRIM7 E3 ligase may represent a new type of Gln/C-end degron pathway for some proteins such as RWDD2B. Additionally, viral or cellular proteases could also cleave cellular proteins, generating new N- or C-ends to enable binding with the recognition elements46,47,48. Therefore, the exposed C-terminal glutamine-end motifs in proteins by protease cleavage may also be recognized by TRIM7.
Methods
Protein expression and purification
DNA encoding TRIM7PRY-SPRY (amino acids 338–511) and CVB3 2CΔN (amino acids 117–329) proteins were synthesized (Genewiz), and cloned into expression vectors (pET15 for TRIM7, pET28-SUMO for 2C). Point mutations and truncations were generated using the PCR site-directed Quick-Change mutagenesis. The resultant plasmids were transformed into competent BL21(DE3) E. coli cells for protein expression. Cells were grown to mid-log phase and 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) was added to induce protein expression. Pelleted cells were resuspended in lysis buffer A (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol) and lysed by sonication. Ni-NTA resin (QIAGEN) was added to the lysate, incubated at 4 °C for 1 h and then washed with buffer A. Protein was eluted with elution buffer (25 mM Tris-HCl pH 8.0, 500 mM NaCl, 300 mM imidazole, 2 mM β-mercaptoethanol). His-tagged TRIM7PRY-SPRY was further purified using a Superdex 200 size exclusion column (Cytiva) with a running buffer containing 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM dithiothreitol (DTT). TEV protease was used to remove the His-SUMO tag for 2CΔN, which was further purified by ion-exchange chromatography using HiTrap Q column (Cytiva). Peak fractions were collected and concentrated for use. We also generated a fusion construct in which the 2C fragment (amino acids 319–329) was C-terminally linked to TRIM7PRY-SPRY (amino acids 338–511) using a (GSA)5 linker. The fusion protein was purified by the same protocol as described above. DNA encoding peptides derived from the C-terminal last 10 residues of the norovirus and cellular proteins were cloned into the pET-His-GST vector (Addgene: 29655) with an N-terminal GST tag. Proteins were purified using the GST affinity chromatography (Cytiva). The eluted proteins were further purified using the HiTrap Q column (Cytiva). Peak fractions were collected and concentrated for the ITC binding experiments. The plasmid encoding full-length NSP5 was a kind gift from J. Lei49. Protein expression and purification were performed similarly as described for 2CΔN. In brief, the cell pellets were lysed in lysis buffer A by sonication. After purification using the Ni-NTA beads, HRV 3C protease was added to the eluate for cleavage. The sample was passed through the Ni-NTA beads to obtain the tag-free target proteins. The eluate was further purified using the Superdex 200 column (Cytiva) pre-equilibrated with the buffer containing 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM DTT.
Crystallization, data collection and structure determination
TRIM7PRY-SPRY was concentrated to ~16 mg ml−1 and incubated with synthesized peptides (Sangon Biotech) on ice for 30 min with a molar ratio of 1:2. The fusion protein was concentrated to ~15 mg ml−1. Crystallization was performed using the sitting-drop vapor diffusion method at 18 °C. The crystals were obtained by mixing 1 μl protein solution and 1 μl crystallization buffer. The TRIM7–2C peptide complex crystals were grown in a reservoir solution containing 100 mM citric acid pH 5.0, 20% (w/v) PEG6000, or 20 mM potassium nitrate, 20% (w/v) PEG3350. The TRIM7–2C fusion protein crystal was grown in a reservoir solution containing 200 mM ammonium acetate, 150 mM magnesium acetate tetrahydrate, 5% (w/v) PEG4000. The TRIM7–NSP5 peptide complex crystals were grown in a reservoir solution containing 1.2 M potassium sodium tartrate tetrahydrate, 100 mM Tris pH 8.0. The TRIM7–NSP8 peptide complex crystals were grown in a reservoir solution containing 2.5 M sodium chloride, 100 mM sodium acetate/acetic acid pH 4.5, 200 mM lithium sulfate. The TRIM7–NSP12 peptide complex crystals were grown in a reservoir solution containing 200 mM dl-malic acid pH 7.0, 20% (w/v) PEG3350.
Crystals were cryoprotected in the mother liquor supplemented with 20% glycerol before being flash frozen in liquid nitrogen. X-ray diffraction datasets were collected at beamline BL18U1 of Shanghai Synchrotron Radiation Facility50. Structures were solved by molecular replacement using the apo TRIM7PRY-SPRY structure (PDB ID: 6UMA) as a searching model in CCP4I2 package51. Structures of bound peptides were manually constructed using the COOT program52. The refinement of structures was performed with PHENIX53,54 and CCP4I2 (ref. 51). Statistics of data processing and structural refinement are listed in Supplementary Tables 1 and 3.
Isothermal titration calorimetry
All the isothermal titration calorimetry (ITC) experiments were carried out in a buffer containing 25 mM Tris-HCl pH 8.0, 150 mM NaCl (unless otherwise stated) using a MicroCal PEAQ-ITC system (Malvern) at 25 °C. A typical titration experiment involved 19 injections of protein or peptide substrate (300 μM to 2 mM) solution into the cell containing TRIM7PRY-SPRY protein (20–50 μM). Data were analyzed using MicroCal PEAQ-ITC analysis software according to the ‘one set of site’ fitting model. All the experiments were repeated at least 2–3 times independently with similar results, and one representative plot with the derived dissociation constant KD and standard error of fitting for each experiment was shown.
Molecular dynamics simulation
To understand the effect of the Q329E mutation on the TRIM7–2C association, MD simulation was performed. Amber FF14SB force field55 was used for the WT and Q329E complexes. Both complexes are solvated in TIP3P water so that the minimum distance between the box boundary and any atom in the complexes is 10 Å. As a result, 11,182 and 11,179 water molecules are added to the complex systems, correspondingly. Both systems follow the same simulation routine. The time step is 2 fs with the SHAKE algorithm56 to fix the hydrogen atoms. The cutoff for calculating the electrostatic interaction is 8 Å. First, 3,000 steps steepest descent followed by 2,000 steps conjugate gradient minimization are performed. After that, additional 7,000 steps steepest descent followed by 3,000 steps conjugate gradient are performed without restraint. After the minimization, the system is heated up to 300 K within 20 ps. The system is then equilibrated in the NPT ensemble for 100 ps, followed by a further equilibration of 100 ps in NPT. The temperature is controlled at 300 K by Langevin dynamics. The pressure is controlled at 1 bar by isotropic Berendsen barostat. Both the temperature and pressure are regulated every 1 ps. The production MD simulation is carried out in a NPT ensemble with the same parameter settings. The cpptraj module was used for the analysis of trajectories57.
Cell culture and transfection
Hemagglutinin (HA)-tagged full-length human TRIM7 was constructed by inserting into the pCMV-HA vector. FLAG-tagged full-length RWDD2B, norovirus NTPase, SARS-CoV-2 NSP5 and NSP8 were cloned into pCMV-Tag2B vector. HEK293T cells were cultured in Dulbecco’s modified eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum, 2 mM glutamine and 100 U ml−1 penicillin–streptomycin at 5% CO2, 37 °C. Transfection was performed with polyethyleneimine (PEI) (Sigma, 919012) unless otherwise described. Cells cultured in six-well plates were prepared in serum-free DMEM medium. For each well, 3 µg DNA and 9 µg PEI were pre-incubated in 200 µl serum-free DMEM by vertexing for 15 min. The PEI/DNA mixture was then added into cells and incubated for 4 h at 37 °C. Serum-free medium with DNA/PEI mixture was then replaced by the complete medium. Cells were then cultured for 24–48 h to express target proteins.
Lentivirus-mediated stable cell line construction
To generate the stable expression cell lines of HA-TRIM7/TRIM7-mut, HEK293T cells were transfected with pLV-EGFP-Control, pCMV-HA-TRIM7/TRIM7-mut, VSV-G, and psPAX2 plasmids to generate lentivirus. The supernatant was collected and concentrated by 4 × PEG8000 solution 48 h after transfection. HEK293T cells were infected with the concentrated virus, supplied with 8 μg ml−1 Polybrene. After 48 h of infection, stable EGFP-positive cells were isolated by fluorescence-activated cell sorting.
TRIM7-knockout using the CRISPR–Cas9 system
To generate a TRIM7-knockout cell line, lentiCRISPRv2 system (Addgene: 52961) containing the single guide RNA target sequence 5′-GGGTCGGCTGCTAGGCCGCC-3′ (Tsingke Biotechnology) was used to transduce HEK293T cells. Three days after 2 μM of puromycin selection, single clones were obtained. TRIM7-knockout clones were identified by immunoblotting with anti-TRIM7 antibody (Bioss, bs-9164R).
Co-immunoprecipitation and immunoblot analysis
For co-immunoprecipitation, 10 μM MG132 (MedChemExpress, HY-13259) was added into culture medium at 24 h post-transfection, and cells were collected at 30 h post-transfection and washed with PBS buffer before being lyzed in the cold lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1% (v/v) NP-40) supplemented with the protease inhibitor cocktail (Roche Applied Science) unless otherwise described. The lysate was cleared by centrifugation at 13,600 g at 4 °C. Then 500 μg of the supernatant was incubated with 2 μg of antibody followed by incubation with 10 μl 50% slurry of Protein A/G agarose (Thermo Fisher Scientific). After extensive washing, the immunoprecipitated proteins were boiled for SDS-PAGE and immunoblot analysis. Primary antibodies, including anti-HA (Bioss, bsm-33003M), anti-FLAG (CST, 14793), anti-Myc (Santa Cruz, sc-40), anti-RWDD2B (Solarbio, K108360P), anti-TRIM7 (Bioss, bs-9164R) and anti-actin (CST, 4967), were used. The assays were performed at least three biological replicates, and the presented data were representative.
Ubiquitination assays
HEK293T cells were co-transfected with HA-TRIM7, Ubi-Myc and the FLAG-tagged plasmid expressing substrate proteins. After that, cells were treated with 10 μM MG132 for 6 h, washed with cold PBS buffer, collected and lyzed with the cold lysis buffer in the presence of protease inhibitor cocktail. The FLAG-tagged substrate was purified by immunoprecipitation using the anti-FLAG antibody (CST, D6W5B). The ubiquitin mutants Ubi-K11R-Myc, Ubi-K48R-Myc and Ubi-K63R-Myc were used to determine the type of ubiquitin chain linkage mediated by TRIM7 in TRIM7−/− cells. Ubiquitinated proteins were purified using the anti-Myc antibody (Santa Cruz, sc-40) and then subjected to western blot analysis.
For the assay using endogenous ubiquitin, HA-TRIM7 or HA-TRIM7-Mut plasmid was transfected to TRIM7−/− HEK293T cells for 24 h. Six hours after treatment with 10 μM MG132, the cells were lyzed in a denaturing lysis buffer (50 mM Tris-HCl pH 8.0, 2% (v/v) SDS, 5 mM DTT, 150 mM NaCl, 0.3% NP-40 and 2 mM EDTA). The lysates were heated to 95 °C for 5 min, cooled to room temperature and diluted (1:10) with co-immunoprecipitation lysis buffer supplemented with protease inhibitor for immunoprecipitation using the anti-FLAG antibody and analyzed with endogenous ubiquitin antibody (CST, 3936T).
Protein stability and degradation assays
HEK293T cells were co-transfected with indicated expression plasmids encoding potential TRIM7 substrates, and increasing amounts of HA-TRIM7/TRIM7-mut plasmid. Eighteen hours after transfection, cells were treated with 10 μM MG132 or DMSO for 6 h. Then, cells were lyzed in the cold lysis buffer with protease inhibitor and protein levels of substrates proteins were assessed by western blot. The experiment for endogenous RWDD2B protein stability was performed as described above. The RWDD2B protein levels in WT, TRIM7−/− and TRIM7OV HEK293T cells were analyzed by western blot using anti-RWDD2B antibody (Solarbio, K108360P). For NSP5 and NSP8, HEK293T cells of different TRIM7 expression statuses were transfected with indicated plasmids, and then the substrates were assessed by western blot. For the cycloheximide chase assay, after transient transfection with the indicated plasmids, HEK293T cells were treated with 10 μg ml−1 of cycloheximide (MedChemExpress, HY-12320), followed by sample collection at indicated time-points (0, 2, 4, 6, 8 and 10 h). Cells were lyzed in cold lysis buffer supplemented with protease inhibitor and the cell extracts were analyzed by western blotting using the indicated antibodies. The presented data were representative of at least three biological replicates.
Dual-luciferase reporter assay
HEK293T cells stably expressing HA-TRIM7/TRIM7-mut or EGFP were co-transfected with 20–80 ng of indicated expression plasmids or empty vector, and luciferase plasmid cocktail containing 10 ng of IFN-β-firefly-luc plasmid (IFN-β promoter-driven firefly luciferase plasmid) and 5 ng of pTracer-Renilla-luc plasmid (luciferase control plasmid). Cells were stimulated by co-transfection with 10 ng of pcDNA3-RIG-I CARD plasmids to induce IFN-β production, and empty pcDNA3 vector was used as the unstimulated control. Lipofectamine 3000 reagent (Invitrogen, L3000001) was used to transfect these plasmids. The cells were assayed for luciferase activities using a dual-luciferase assay kit (Beyotime, RG088S) 24 h after transfection according to the manufacturer’s instructions. The assays were performed with at least three biological replicates, and the presented data were representative.
Enzyme-linked immunosorbent assay
HEK293T cells were transfected with the indicated plasmids for 12 h, and then infected with VSV (multiplicity of infection of 0.1) for 12 h. The amount of secreted IFN-β in the supernatant was then measured using a human IFN-β ELISA kit (Multi Sciences, EK1236) following the manufacturer’s instructions. The assays were performed on at least three biological replicates.
Analysis of phosphorylation
HEK293T cells were co-transfected with TBK1-Myc-expressing plasmid, NSP5/NSP8-encoding plasmids and HA-TRIM7 expression plasmids. Western blotting was used to analyze the cell lysates for S172-phosphorylated TBK1 (CST, 5483), total TBK1 (CST, 3013S), and indicated proteins 24 h after transfection. The presented data were representative of at least three biological replicates.
Statistics analysis
All the statistical data were analyzed and plotted with Graphpad Prism 8. Statistical significance was determined using two-way ANOVA or one-way ANOVA with Tukey’s correction. Error bars represent mean ± s.e.m. of biological triplicates. P values were shown by n.s. (no significant) or stars (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The structure factors and atomic coordinates for TRIM7 in complex with substrate peptides have been deposited to the Protein Data Bank with accession numbers of 7W0Q (TRIM7–2C, 1.1 Å), 7W0S (TRIM7–2C, 1.4 Å), 7W0T (TRIM7–2C, 1.57 Å), 7X6Y (TRIM7–NSP5, 1.39 Å), 7X70 (TRIM7–NSP8, 1.25 Å) and 7X6Z (TRIM7–NSP12, 1.43 Å). Source data are provided with this paper.
References
Hatakeyama, S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 42, 297–311 (2017).
Reymond, A. et al. The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001).
Shen, Z. et al. The roles of TRIMs in antiviral innate immune signaling. Front. Cell. Infect. Microbiol. 11, 628275 (2021).
Giraldo, M. I., Hage, A., van Tol, S. & Rajsbaum, R. TRIM proteins in host defense and viral pathogenesis. Curr. Clin. Microbiol. Rep. 7, 101–114 (2020).
Koepke, L., Gack, M. U. & Sparrer, K. M. The antiviral activities of TRIM proteins. Curr. Opin. Microbiol. 59, 50–57 (2021).
Hage, A. & Rajsbaum, R. To TRIM or not to TRIM: the balance of host–virus interactions mediated by the ubiquitin system. J. Gen. Virol. 100, 1641 (2019).
van Gent, M., Sparrer, K. M. & Gack, M. U. TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol. 5, 385–405 (2018).
D’Cruz, A. A., Babon, J. J., Norton, R. S., Nicola, N. A. & Nicholson, S. E. Structure and function of the SPRY/B30. 2 domain proteins involved in innate immunity. Protein Sci. 22, 1–10 (2013).
Montori-Grau, M. et al. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism 83, 177–187 (2018).
Chakraborty, A., Diefenbacher, M. E., Mylona, A., Kassel, O. & Behrens, A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat. Commun. 6, 1–12 (2015).
Hu, X. et al. Tripartite motif-containing protein 7 regulates hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway. Biochem. Biophys. Res. Commun. 511, 889–895 (2019).
Fan, W. et al. TRIM7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity. Cell 184, 3410–3425 (2021).
Giraldo, M. I. et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature 585, 414–419 (2020).
Orchard, R. C. et al. Identification of antinorovirus genes in human cells using genome-wide CRISPR activation screening. J. Virol. 93, e01324–01318 (2019).
Yang, B. et al. RNF90 negatively regulates cellular antiviral responses by targeting MITA for degradation. PLoS Pathog. 16, e1008387 (2020).
Yang, B. et al. Negative regulation of RNF90 on RNA virus-triggered antiviral immune responses targeting MAVS. Front. Immunol. 12, 730483 (2021).
Sosa, C. J. M., Issoglio, F. M. & Carrizo, M. E. Crystal structure and mutational analysis of the human TRIM7 B30. 2 domain provide insights into the molecular basis of its binding to glycogenin-1. J. Biol. Chem. 296, 100772 (2021).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Zheng, Z. et al. Human norovirus NTPase antagonizes interferon-β production by interacting with IkB kinase ε. Front. Microbiol. 12, 687933 (2021).
Xia, H. et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020).
Liu, Y. et al. SARS-CoV-2 Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. MBio 12, e02335–02321 (2021).
Zheng, Y. et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 7, 1–12 (2022).
Shemesh, M. et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 17, e1009800 (2021).
Yang, Z. et al. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. Preprint at https://doi.org/10.1101/2020.08.12.247767 (2020).
Banerjee, A. K. et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183, 1325–1339 (2020).
Zhou, C. et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine 59, 102955 (2020).
Zhai, L., Dietrich, A., Skurat, A. V. & Roach, P. J. Structure–function analysis of GNIP, the glycogenin-interacting protein. Arch. Biochem. Biophys. 421, 236–242 (2004).
Wang, Z. et al. Tripartite motif containing 11 interacts with DUSP6 to promote the growth of human osteosarcoma cells through regulating ERK1/2 pathway. BioMed Res. Int. 25, 9612125 (2019).
Luck, K. et al. A reference map of the human binary protein interactome. Nature 580, 402–408 (2020).
Campillay-Véliz, C. P. et al. Human norovirus proteins: implications in the replicative cycle, pathogenesis, and the host immune response. Front. Immunol. 11, 961 (2020).
Mariano, G., Farthing, R. J., Lale-Farjat, S. L. & Bergeron, J. R. Structural characterization of SARS-CoV-2: where we are, and where we need to be. Front. Mol. Biosci. 7, 344 (2020).
Chen, J. et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 182, 1560–1573. e1513 (2020).
Kneller, D. W. et al. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 11, 1–6 (2020).
Tan, J. et al. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 87, 4339–4351 (2013).
Sosnovtsev, S. V. et al. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 80, 7816–7831 (2006).
Ziebuhr, J., Heusipp, G. & Siddell, S. G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 71, 3992–3997 (1997).
Parker, E. et al. Multi‐tissue epigenetic and gene expression analysis combined with epigenome modulation identifies RWDD2B as a target of osteoarthritis susceptibility. Arthritis Rheumatol. 73, 100–109 (2021).
Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl Acad. Sci. 116, 358–366 (2019).
Koren, I. et al. The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell 173, 1622–1635 (2018).
Lin, H.-C. et al. C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol. cell 70, 602–613. e603 (2018).
Chen, X. et al. Molecular basis for arginine C-terminal degron recognition by Cul2FEM1 E3 ligase. Nat. Chem. Biol. 17, 254–262 (2021).
Yan, X. et al. Molecular basis for ubiquitin ligase CRL2FEM1C-mediated recognition of C-degron. Nat. Chem. Biol. 17, 263–271 (2021).
Rusnac, D.-V. et al. Recognition of the diglycine C-end degron by CRL2KLHDC2 ubiquitin ligase. Mol. cell 72, 813–822. e814 (2018).
Li, Y. et al. Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase. Proc. Natl Acad. Sci. 115, 319–324 (2018).
Zhao, S. et al. Structural insights into SMCR8 C-degron recognition by FEM1B. Biochem. Biophys. Res. Commun. 557, 236–239 (2021).
Tsu, B. V. et al. Diverse viral proteases activate the NLRP1 inflammasome. eLife 10, e60609 (2021).
Saita, S. et al. PARL mediates Smac proteolytic maturation in mitochondria to promote apoptosis. Nat. Cell Biol. 19, 318–328 (2017).
Robinson, K. S. et al. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, eaay2002 (2020).
Qiao, J. et al. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science 371, 1374–1378 (2021).
Zhang, W.-Z. et al. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 30, 1–11 (2019).
Potterton, L. et al. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. Sect. D Struct. Biol. 74, 68–84 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr., Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 1948–1954 (2002).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 68, 352–367 (2012).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Roe, D. R. & Cheatham, T. E. III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Acknowledgements
We thank J. Lei for supporting plasmids encoding SARS-CoV-2 proteins. We thank the staffs from BL17B/BL18U1/BL19U1/BL19U2/BL01B beamlines of the National Facility for Protein Science in Shanghai at Shanghai Synchrotron Radiation Facility, for assistance during data collection. This work is supported by the National Natural Science Foundation of China Grants (32071218 to H.Z.) and the National Key Research and Development Program of China (2018YFC1313002 to L.L.).
Author information
Authors and Affiliations
Contributions
X.L., Y.W., Y.M. and Y.Z. performed the sample preparation, biochemistry and crystallization experiments. Xu.L. collected the diffraction data and solved the structure with the help from X.L. and Y. M. X.C. performed the molecular dynamics analysis. J.X., Y. Li., Y. Lu, X.L., Z.L., X.Z., D.J. and P.W. carried out the cellular experiments. C.X., L.L., G.Y. and H.Z. analyzed the data. H.Z., G.Y. and L.L. designed the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
X.C. is the founder of YDS Pharmatech. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Chao Xu, Matthew Ravalin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 TRIM7 interacts with enteroviruses 2C.
a, Sequence alignment of 2C proteins from different enteroviruses. The last eight residues at the C-terminus are displayed. Invariant and conserved residues are shaded green and yellow, respectively. b, ITC measurements of peptides derived from the C-terminal fragments of 2C proteins from different enteroviruses, including enterovirus 71, poliovirus and enterovirus D68, to TRIM7PRY-SPRY.
Extended Data Fig. 2 Structural features of the TRIM7PRY-SPRY-2C peptide complex.
a, The overall structures of TRIM7PRY-SPRY-2C peptide complex solved in different space groups (top). Bottom, 2Fo-Fc electron density map of the 2C fragments contoured at 1.0 σ. b, Structural comparison of the three TRIM7PRY-SPRY-2C peptide structures and a previously published substrate-free structure (PDB ID: 6UMA). c, ITC measurement of TRIM7PRY-SPRY-2CΔN association in a high salt buffer containing 500 mM NaCl. d, Conservation scores across different species were mapped to the TRIM7 structure. Multiple sequence alignment of human TRIM7 and orthologs in other mammals, birds and amphibians were performed with Clustal Omega. The multiple sequence alignment and the resolved structure of TRIM7PRY-SPRY were used as inputs for conservation analysis using the ConSurf web server (https://consurf.tau.ac.il/).
Extended Data Fig. 3 Quantification of the binding affinity between TRIM7 and CVB3 2C mutations.
Thermodynamic analysis of the interaction between TRIM7PRY-SPRY and 2C mutants (T323G, T323S, T323I).
Extended Data Fig. 4 MD simulations and ITC measurements of the interaction between TRIM7PRY-SPRY and 2C variants.
a, (left) Structural comparison between TRIM2-2C and the calculated TRIM7-2C (Q329E). (right) Effects of simulations on the Q329E mutation. Molecular dynamics simulation suggested that Q329E mutation eliminates the interactions between the residue 329 and TRIM7, especially the interactions with Arg385, Asn438, and Ser499 in TRIM7 (Supplementary Table 2). Consequently, the C-terminal carboxylate group of 2C at residue 329 is more exposed to solvent, leading to a further reduced TRIM7-2C interaction and increased 2C C-terminal solvation. b, The root-mean-square deviation (RMSD) change of the 2C peptide structures in MD simulation (left). The RMSD values of WT (black) and Q329E (red) 2C peptide structures with reference to the first frame of the trajectory in MD simulation were calculated and plotted. While the WT 2C peptide gradually converged to the original position, the Q329E mutant drifted away as indicated by the increasing RMSD over MD simulation. Right, the Q329E mutation led to increased hydrogen bonding with water. The number of hydrogen-bonded water molecules to the side chain amide of Q329 in WT 2C (black) and the side chain carboxyl of E329 in the mutant (red) were plotted over MD simulation.
Extended Data Fig. 5 NSP proteins from SARS-CoV-2 counteract the IFNB1 promoter activity.
a, Co-immunoprecipitation of TRIM7 with NSP5 and NSP8. FLAG-tagged NSP5, NSP8 or empty vector was co-transfected with HA-TRIM7 in HEK293T cells. Cells were lysed and immunoprecipitated with anti-HA antibody and detected by anti-FLAG antibody. HA-TRIM7-Mut, TRIM7 catalytic dead mutant (C29A/C31A); NSP5/NSP8-Q/A, the alanine replacement of the C-terminal glutamine in NSP5/8. b, Crystal structure of TRIM7PRY-SPRY in complex with NSP12 peptide (aa 928-932). c, 2Fo-Fc electron density map of the TRIM7-bound NSP peptides contoured at 1.0 σ. d, Luciferase activity of IFNB1 promoter reporter in HEK293T cells transfected with indicated NSP proteins and RIG-I CARD expression plasmids. Relative luciferase activity was quantified 24 h post-transfection. Data are presented as mean ± SEM; n = 6, biologically independent samples were examined over three independent experiments. Statistical significance was determined by comparing with ‘Vector’ group using one-way ANOVA with Tukey’s correction. e, f, Analysis of TBK1 phosphorylation in the presence of NSP5 (e) or NSP8 (f).
Extended Data Fig. 6 ITC measurements of TRIM7PRY-SPRY with multiple cellular proteins functionally linked to TRIM7.
a, TRIM7PRY-SPRY binds with GN1 and RACO-1 C-terminus peptide fragments. b, Thermodynamic analysis of the interaction between TRIM7PRY-SPRY and BRMS1 peptide (left panel), DUSP6 peptide (middle panel) and STING protein (right panel).
Supplementary information
Supplementary Information
Supplementary Figs 1–11, Supplementary Tables 1–3 and uncropped gel images.
Source data
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, X., Xiao, J., Li, X. et al. A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures. Nat Chem Biol 18, 1214–1223 (2022). https://doi.org/10.1038/s41589-022-01128-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01128-x
This article is cited by
-
TRIM-away via Gln/C-degrons
Nature Chemical Biology (2022)