Abstract
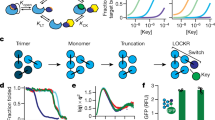
Splitting bioactive proteins into conditionally reconstituting fragments is a powerful strategy for building tools to study and control biological systems. However, split proteins often exhibit a high propensity to reconstitute, even without the conditional trigger, limiting their utility. Current approaches for tuning reconstitution propensity are laborious, context-specific or often ineffective. Here, we report a computational design strategy grounded in fundamental protein biophysics to guide experimental evaluation of a sparse set of mutants to identify an optimal functional window. We hypothesized that testing a limited set of mutants would direct subsequent mutagenesis efforts by predicting desirable mutant combinations from a vast mutational landscape. This strategy varies the degree of interfacial destabilization while preserving stability and catalytic activity. We validate our method by solving two distinct split protein design challenges, generating both design and mechanistic insights. This new technology will streamline the generation and use of split protein systems for diverse applications.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request. Raw experimental data and computation generated data for main text figures are provided as Source data. Raw experimental data for Supplementary figures are provided in Supplementary Data 1. Plasmid maps are provided in Supplementary Data 2, and annotated descriptions of all plasmids are in Supplementary Data 3. The structure of TEVp was obtained from the Research Crystallography for Structural Bioinformatics (RCSB) (PDB: 1LVM). A subset of plasmids used in this study will be made available on Addgene, including complete and annotated GenBank files, at https://www.addgene.org/Joshua_Leonard/. Source data are provided with this paper.
References
Romei, M. G. & Boxer, S. G. Split green fluorescent proteins: scope, limitations and outlook. Annu. Rev. Biophys. 48, 19–44 (2019).
Shekhawat, S. S. & Ghosh, I. Split-protein systems: beyond binary protein–protein interactions. Curr. Opin. Chem. Biol. 15, 789–797 (2011).
Wehr, M. C. & Rossner, M. J. Split protein biosensor assays in molecular pharmacological studies. Drug Discov. Today 21, 415–429 (2016).
Muller, J. & Johnsson, N. Split-ubiquitin and the split-protein sensors: chessman for the endgame. ChemBioChem 9, 2029–2038 (2008).
Paulmurugan, R. & Gambhir, S. S. Monitoring protein–protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 75, 1584–1589 (2003).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Ozawa, T., Kaihara, A., Sato, M., Tachihara, K. & Umezawa, Y. Split luciferase as an optical probe for detecting protein–protein interactions in mammalian cells based on protein splicing. Anal. Chem. 73, 2516–2521 (2001).
Gray, D. C., Mahrus, S. & Wells, J. A. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 142, 637–646 (2010).
Gao, X. J., Chong, L. S., Kim, M. S. & Elowitz, M. B. Programmable protein circuits in living cells. Science 361, 1252–1258 (2018).
Fink, T. et al. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat. Chem. Biol. 15, 115–122 (2019).
Zetsche, B., Volz, S. E. & Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015).
Nihongaki, Y., Otabe, T., Ueda, Y. & Sato, M. A split CRISPR–Cpf1 platform for inducible genome editing and gene activation. Nat. Chem. Biol. 15, 882–888 (2019).
Paulmurugan, R., Umezawa, Y. & Gambhir, S. S. Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl Acad. Sci. USA 99, 15608–15613 (2002).
Fetchko, M. & Stagljar, I. Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32, 349–362 (2004).
Pandey, N., Nobles, C. L., Zechiedrich, L., Maresso, A. W. & Silberg, J. J. Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein–protein interactions. ACS Synth. Biol. 4, 615–624 (2015).
Jones, K. A. et al. Development of a split esterase for protein–protein interaction-dependent small-molecule activation. ACS Cent. Sci. 5, 1768–1776 (2019).
Wehr, M. C., Reinecke, L., Botvinnik, A. & Rossner, M. J. Analysis of transient phosphorylation-dependent protein–protein interactions in living mammalian cells using split-TEV. BMC Biotechnol. 8, 55 (2008).
Camacho-Soto, K., Castillo-Montoya, J., Tye, B. & Ghosh, I. Ligand-gated split-kinases. J. Am. Chem. Soc. 136, 3995–4002 (2014).
Camacho-Soto, K., Castillo-Montoya, J., Tye, B., Ogunleye, L. O. & Ghosh, I. Small molecule gated split-tyrosine phosphatases and orthogonal split-tyrosine kinases. J. Am. Chem. Soc. 136, 17078–17086 (2014).
Dagliyan, O. et al. Computational design of chemogenetic and optogenetic split proteins. Nat. Commun. 9, 4042 (2018).
Silberg, J. J., Endelman, J. B. & Arnold, F. H. SCHEMA-guided protein recombination. Methods Enzymol. 388, 35–42 (2004).
Nguyen, P. Q., Liu, S., Thompson, J. C. & Silberg, J. J. Thermostability promotes the cooperative function of split adenylate kinases. Protein Eng. Des. Selection 21, 303–310 (2008).
Lindman, S., Hernandez-Garcia, A., Szczepankiewicz, O., Frohm, B. & Linse, S. In vivo protein stabilization based on fragment complementation and a split GFP system. Proc. Natl Acad. Sci. USA 107, 19826–19831 (2010).
Dantas, G. et al. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J. Mol. Biol. 366, 1209–1221 (2007).
Yin, S., Ding, F. & Dokholyan, N. V. Eris: an automated estimator of protein stability. Nat. Methods 4, 466–467 (2007).
Masso, M. & Vaisman, I. I. Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics 24, 2002–2009 (2008).
Lee, T.-S. & York, D. M. Computational mutagenesis studies of hammerhead ribozyme catalysis. J. Am. Chem. Soc. 132, 13505–13518 (2010).
Han, Y. et al. Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635 (2019).
Daringer, N. M., Dudek, R. M., Schwarz, K. A. & Leonard, J. N. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol. 3, 892–902 (2014).
Wehr, M. C. et al. Monitoring regulated protein–protein interactions using split TEV. Nat. Methods 3, 985–993 (2006).
Crescitelli, R. et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles https://doi.org/10.3402/jev.v2i0.20677 (2013).
Yen, H.-C. S., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008).
Edelstein, H. I. et al. Elucidation and refinement of synthetic receptor mechanisms. Synth. Biol. (2020); https://doi.org/10.1093/synbio/ysaa017
Kapust, R. B., Tozser, J., Copeland, T. D. & Waugh, D. S. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 294, 949–955 (2002).
Hartfield, R. M., Schwarz, K. A., Muldoon, J. J., Bagheri, N. & Leonard, J. N. Multiplexing engineered receptors for multiparametric evaluation of environmental ligands. ACS Synth. Biol. 6, 2042–2055 (2017).
Schwarz, K. A., Daringer, N. M., Dolberg, T. B. & Leonard, J. N. Rewiring human cellular input–output using modular extracellular sensors. Nat. Chem. Biol. 13, 202–209 (2017).
Donahue, P. S. et al. The COMET toolkit for composing customizable genetic programs in mammalian cells. Nat. Commun. 11, 779 (2020).
Xia, Z. & Liu, Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81, 2395–2402 (2001).
Eisenhaber, F., Lijnzaad, P., Argos, P., Sander, C. & Scharf, M. The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J. Comput. Chem. 16, 273–284 (1995).
Alford, R. F. et al. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 3031–3048 (2017).
Acknowledgements
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering of the NIH under award no. 1R01EB026510 (J.N.L.) and the Northwestern University Flow Cytometry Core Facility supported by a Cancer Center Support Grant (NCI 5P30CA060553). T.B.D was supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG). J.D.B. and A.N.P. were supported by the National Science Foundation through Graduate Research Fellowships. J.D.B. and W.K.C. were supported in part by the National Institutes of Health Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. This work is also supported in part by the Great Lakes Bioenergy Research Center, US Department of Energy, Office of Science, Office of Biological and Environmental Research, under award no. DE-SC0018409 (S.R. and A.T.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, Department of Defense, Department of Energy or other federal agencies.
Author information
Authors and Affiliations
Contributions
T.B.D., A.T.M., S.R. and J.N.L. conceptualized the project. T.B.D., J.D.B., W.K.C. and E.E.S. created reagents, designed and performed experiments, and analyzed the data. A.N.P. assisted in analyzing and visualizing the data. A.T.M. developed the computational model and code. T.B.D., A.T.M., S.R. and J.N.L. drafted the manuscript. T.B.D., A.T.M. and A.N.P. created the figures. J.N.L. and S.R. supervised the work. All authors edited and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.N.L. is a co-inventor on a patent that covers the MESA technology used in this manuscript (US patent 9,732,392 B2). The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14, Tables 1–4, Note and references.
Supplementary Data 1
Raw data for Supplementary figures.
Supplementary Software 1
Linear discriminate analysis script. Note that this is a text file with a file extension indicating that it is for use with R.
Supplementary Software 2
Jupyter notebook code for Fig. 4a. Note that this is a text file with a file extension indicating that it is for use with Jupyter.
Supplementary Data 2
Archive of plasmid maps.
Supplementary Data 3
List of all plasmids and annotation of key features.
Source data
Source Data Fig. 2
Raw data.
Source Data Fig. 3
Raw data.
Source Data Fig. 4
Raw data.
Source Data Fig. 5
Raw data.
Rights and permissions
About this article
Cite this article
Dolberg, T.B., Meger, A.T., Boucher, J.D. et al. Computation-guided optimization of split protein systems. Nat Chem Biol 17, 531–539 (2021). https://doi.org/10.1038/s41589-020-00729-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-00729-8
This article is cited by
-
Understanding blaNDM-1 gene regulation in CRKP infections: toward novel antimicrobial strategies for hospital-acquired pneumonia
Molecular Medicine (2024)
-
Irreversible light-activated SpyLigation mediates split-protein assembly in 4D
Nature Protocols (2024)
-
Programmable synthetic receptors: the next-generation of cell and gene therapies
Signal Transduction and Targeted Therapy (2024)
-
Integrated compact regulators of protein activity enable control of signaling pathways and genome-editing in vivo
Cell Discovery (2024)
-
A single-component, light-assisted uncaging switch for endoproteolytic release
Nature Chemical Biology (2024)