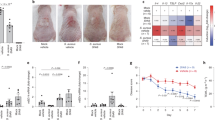
Abstract
Microorganisms can be equipped with synthetic genetic programs for the production of targeted therapeutic molecules. Cutibacterium acnes is the most abundant commensal of the human skin, making it an attractive chassis to create skin-delivered therapeutics. Here, we report the engineering of this bacterium to produce and secrete the therapeutic molecule neutrophil gelatinase-associated lipocalin, in vivo, for the modulation of cutaneous sebum production.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Sequencing datasets generated during this study are available from the NCBI Sequence Read Archive (SRA) with BioProject ID number PRJNA1007560 (ref. 63). The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE64 partner repository with the dataset identifier PXD044802 (ref. 65). Source data are provided with this paper.
References
Leventhal, D. S. et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 11, 2739 (2020).
Isabella, V. M. et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018).
Mao, N., Cubillos-Ruiz, A., Cameron, D. E. & Collins, J. J. Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 10, eaao2586 (2018).
Danino, T. et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7, 289ra84 (2015).
Hwang, I. Y. et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8, 15028 (2017).
Kurtz, C. B. et al. An engineered Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 11, eaau7975 (2019).
Robert, S. et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes 63, 2876–2887 (2014).
Ma, J. et al. Engineered probiotics. Microb. Cell Fact. 21, 72 (2022).
Zhou, Z. et al. Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact. 19, 56 (2020).
Chen, Y. E. et al. Engineered skin bacteria induce antitumor T cell responses against melanoma. Science 380, 203–210 (2023).
Öhnstedt, E. et al. Engineered bacteria to accelerate wound healing: an adaptive, randomised, double-blind, placebo-controlled, first-in-human phase 1 trial. EClinicalMedicine 60, 102014 (2023).
Maura, D., Elmekki, N. & Goddard, C. A. The ammonia oxidizing bacterium Nitrosomonas eutropha blocks T helper 2 cell polarization via the anti-inflammatory cytokine IL-10. Sci. Rep. 11, 14162 (2021).
Lee, N. Y. et al. Dermal microflora restoration with ammonia-oxidizing bacteria Nitrosomonas eutropha in the treatment of keratosis pilaris: a randomized clinical trial. J. Drugs Dermatol. 17, 285–288 (2018).
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 (2018).
Boxberger, M., Cenizo, V., Cassir, N. & La Scola, B. Challenges in exploring and manipulating the human skin microbiome. Microbiome 9, 1–14 (2021).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190 (2009).
Conwill, A. et al. Anatomy promotes neutral coexistence of strains in the human skin microbiome. Cell Host Microbe 30, 171–182 (2022).
Zouboulis, C. C.Acne and sebaceous gland function. Clin. Dermatol. 22, 360–366 (2004).
Gribbon, E. M., Cunliffe, W. J. & Holland, K. T. Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 139, 1745–1751 (1993).
Fournière, M., Latire, T., Souak, D., Feuilloley, M. G. J. & Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 8, 1752 (2020).
Paetzold, B. et al. Skin microbiome modulation induced by probiotic solutions. Microbiome 7, 95 (2019).
Oh, J., Byrd, A. L., Park, M., Kong, H. H. & Segre, J. A.Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016).
Rosenthal, M., Goldberg, D., Aiello, A., Larson, E. & Foxman, B.Skin microbiota: microbial community structure and its potential association with health and disease. Infect. Genet. and Evol. 11, 839–848 (2011).
Dréno, B. et al. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 32, 5–14 (2018).
Moradi Tuchayi, S. et al. Acne vulgaris. Nat. Rev. Dis. Primers 1, 15029 (2015).
Layton, A. The use of isotretinoin in acne. Dermatoendocrinol. 1, 162 (2009).
Nelson, A. M. et al. Temporal changes in gene expression in the skin of patients treated with isotretinoin provide insight into its mechanism of action. Dermatoendocrinol. 1, 177 (2009).
Nelson, A. M. et al. Neutrophil gelatinase–associated lipocalin mediates 13-cis retinoic acid–induced apoptosis of human sebaceous gland cells. J. Clin. Invest. 118, 1468 (2008).
Lumsden, K. R. et al. Isotretinoin increases skin-surface levels of neutrophil gelatinase-associated lipocalin in patients treated for severe acne. Br. J. Dermatol. 165, 302–310 (2011).
Novickii, V. et al. Different permeabilization patterns of splenocytes and thymocytes to combination of pulsed electric and magnetic field treatments. Bioelectrochemistry 122, 183–190 (2018).
Knödlseder, N. et al. Engineering selectivity of Cutibacterium acnes phages by epigenetic imprinting. PLoS Pathog. 18, e1010420 (2022).
Deptula, P. et al. Complete genome sequences and methylome analyses of Cutibacterium acnes subsp. acnes strains DSM 16379 and DSM 1897T. Microbiol. Resour. Announc. 9, e00705–e00720 (2020).
Johnson, B. H. & Hecht, M. H.Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Nat. Biotechnol. 12, 1357–1360 (1994).
Sharma, S. et al. A simple and cost-effective freeze-thaw based method for Plasmodium DNA extraction from dried blood spot. Iran. J. Parasitol. 14, 29 (2019).
Harju, S., Fedosyuk, H. & Peterson, K. R. Rapid isolation of yeast genomic DNA: Bust n’ Grab. BMC Biotechnol. 4, 8 (2004).
Park, S. F. & Stewart, G. S. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94, 129–132 (1990).
Pyne, M. E., Moo-Young, M., Chung, D. A. & Chou, C. P. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol. Biofuels 6, 50 (2013).
Sörensen, M. et al. Mutagenesis of Propionibacterium acnes and analysis of two CAMP factor knock-out mutants. J. Microbiol. Methods 83, 211–216 (2010).
Nazipi, S., Stødkilde, K., Scavenius, C. & Brüggemann, H.The skin bacterium Propionibacterium acnes employs two variants of hyaluronate lyase with distinct properties. Microorganisms 5, 57 (2017).
Allhorn, M., Arve, S., Brüggemann, H. & Lood, R. A. A novel enzyme with antioxidant capacity produced by the ubiquitous skin colonizer Propionibacterium acnes. Sci. Rep. https://doi.org/10.1038/srep36412 (2016).
Pedrolli, D. B. et al. Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 37, 100–115 (2019).
Amalaradjou, M. A. R. & Bhunia, A. K. Bioengineered probiotics, a strategic approach to control enteric infections. Bioengineered 4, 379–387 (2013).
Charbonneau, M. R., Isabella, V. M., Li, N. & Kurtz, C. B. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 11, 1738 (2020).
Norville, J.E. et al. Assembly of radically recoded E. coli genome segments. Preprint at bioRxiv https://doi.org/10.1101/070417 (2016).
Holland, C. et al. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 10, 230 (2010).
Yu, Y., Champer, J. & Kim, J.Analysis of the surface, secreted, and intracellular proteome of Propionibacterium acnes. EuPA Open Proteom. 9, 1–7 (2015).
Lumsden, K. R. The Innate Immune Protein Neutrophil Gelatinase-Associated Lipocalin is Involved in the Early Therapeutic Response to 13-cis Retinoic Acid in Acne Patients. Dissertation, Pennsylvania State University (2009).
Wagner, E. F., Schonthaler, H. B., Guinea-Viniegra, J. & Tschachler, E. Psoriasis: what we have learned from mouse models. Nat. Rev. Rheumatol. 6, 704–714 (2010).
Zomer, H. D. & Trentin, A. G. Skin wound healing in humans and mice: challenges in translational research. J. Dermatol. Sci. 90, 3–12 (2018).
Niehues, H. et al. 3D skin models for 3R research: the potential of 3D reconstructed skin models to study skin barrier function. Exp. Dermatol. 27, 501–511 (2018).
Emmert, H., Rademacher, F., Gläser, R. & Harder, J. Skin microbiota analysis in human 3D skin models—‘Free your mice’. Exp. Dermatol. 29, 1133–1139 (2020).
Jore, J. P., van Luijk, N., Luiten, R. G., van der Werf, M. J. & Pouwels, P. H. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67, 499–503 (2001).
Lood, R. Propionibacterium acnes and its phages. Ph.D. thesis, Lund University (2011).
Kay, M. A., He, C.-Y. & Chen, Z.-Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 28, 1287–1289 (2010).
Johnston, C. D. et al. Systematic evasion of the restriction-modification barrier in bacteria. Proc. Natl Acad. Sci. USA 116, 11454–11459 (2019).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Koontz, L. TCA precipitation. Methods Enzymol. 541, 3–10 (2014).
Zouboulis, C. C., Seltmann, H., Neitzel, H. & Orfanos, C. E. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J. Invest. Dermatol. 113, 1011–1020 (1999).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Knödlseder, N. et al. Mouse skin microbiome samples before and after C. acnes application. NCBI Bioproject, PRJNA1007560. Metagenomics. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1007560 (2023).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Knödlseder, N. et al. Engineered skin microbiome-assisted delivery to the pilosebaceous unit. ProteomeXchange, PXD044802. Proteomics. https://www.ebi.ac.uk/pride/archive/projects/PXD044802 (2023).
Acknowledgements
We want to thank C.D. Johnston from the Fred Hutchinson Cancer Center for kindly providing the JMC2 and JMC3 MC producer strains. We would like to thank T.F. Meyer for the opportunity to visit his lab and M. Sörensen for her technical support and time during the research stay. Furthermore, we want to thank B. Paetzold for the interesting discussions. We want to express our gratitude to E. Sabido from the UPF/CRG proteomics unit for proteomic analysis support. We thank the entire Synbio team for their support and helpful discussions. N.K. thanks A. Rahmeh, M. Pallares and M. Pol for useful discussions and input.
This work was funded by the Office of Naval Research (award N62909-18-1-2155), INNOValora (INNOV21-09-1; SynFlora) given by Universitat Pompeu Fabra and Indústria del Coneixement of the Catalan government (AGAUR; IdC 2019 PROD 00057) and SKINDEV ‘Skin microbial devices’ by European Innovation Council (101098826), all granted to M.G., as well as by research grants PID2020-114477RB-I00 to C.S. and PID2021-126249OA-I00 to J.M. from the MCIN/AEI/10.13039/501100011033. N.K. is funded by a Maria Maetzu–UPF fellowship (AEI–MM-CEX2018-000792-M D.COMAS–Programa estatal de fomento investigación científica y técnica de exceléncia, unidades de exceléncia María de Maeztu 2018) and by the Sociedad Española de Químicos Cosméticos (Beca de la SEQC para la presentación de trabajos en el 33rd IFSCC Congress; KNÖDLSEDER, NASTASSIA). M.J.F. is funded by a Juan de la Cierva Fellowship from the Spanish government (award FJC 2018-037096-I). N.K. also received an EMBO short-term fellowship (fellowship no. 8240). J.S.M. is funded by a Marie Skłodowska-Curie Individual Fellowship (European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie grant agreement 882387), a Juan de la Cierva Incorporación Fellowship (IJC2020-043621-I/MCIN/AEI/10.13039/501100011033 and NextGenerationEU/PRTR from the European Union) and a DCEXS MM Postdoc Project (Unidad de Excelencia María de Maeztu, funded by the AEI, ref. CEX2018-000792-M).
The graphic illustrations in this study were generated using BioRender.com. Data plots were generated using GraphPad Prism 9.
Author information
Authors and Affiliations
Contributions
N.K. conceptualized, designed and carried out the experiments with the guidance of M.G. M.J.F., L.T. and K.B. contributed to the cell culture work. M.M., L.T. and J.S.M. contributed to the bacterial work. J.S.M. with the help of N.K. designed and created the IIIB methylase-proficient E. coli strain. N.K., J. Manils and C.F. performed the mouse experiments and processed the samples. C.C.Z. provided the SZ95 cell line and guidance. J. Maruotti and H.L. provided the Pci-SEB_Cau cell lines, conducted the initial experiments and provided guidance. H.B. analyzed the 16S datasets and contributed with experimental design and advice. R.L. provided support and guidance for the in vivo production of proteins and acquisition of samples. C.C. and K.B. purified the proteins used in this study. C.S. contributed with advice and guidance. N.K., M.G., M.J.F., J.S.M. and J. Manils drafted the initial manuscript. All authors read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
M.G., N.K., M.J.F. and J.S.M. are inventors of a European patent application submitted by University Pompeu Fabra.
Peer review
Peer review information
Nature Biotechnology thanks Peter Larson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Fig. 1–4, Table 1 and unmodified blots
Supplementary Data
Source data for supplementary figures.
Source data
Source Data Figs. 1 and 2
Statistical source data for main figures.
Source Data Fig. 1f
Unprocessed western blot.
Source Data Fig. 1g
Unprocessed western blot.
Source Data Fig. 2d
Unprocessed microscope images.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Knödlseder, N., Fábrega, MJ., Santos-Moreno, J. et al. Delivery of a sebum modulator by an engineered skin microbe in mice. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-023-02072-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-02072-4
This article is cited by
-
Cutibacterium improves skin condition
Nature Reviews Microbiology (2024)