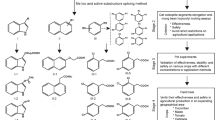
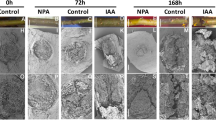
Abstract
Clonal propagation of plants by induction of adventitious roots (ARs) from stem cuttings is a requisite step in breeding programs. A major barrier exists for propagating valuable plants that naturally have low capacity to form ARs. Due to the central role of auxin in organogenesis, indole-3-butyric acid is often used as part of commercial rooting mixtures, yet many recalcitrant plants do not form ARs in response to this treatment. Here we describe the synthesis and screening of a focused library of synthetic auxin conjugates in Eucalyptus grandis cuttings and identify 4-chlorophenoxyacetic acid–l-tryptophan-OMe as a competent enhancer of adventitious rooting in a number of recalcitrant woody plants, including apple and argan. Comprehensive metabolic and functional analyses reveal that this activity is engendered by prolonged auxin signaling due to initial fast uptake and slow release and clearance of the free auxin 4-chlorophenoxyacetic acid. This work highlights the utility of a slow-release strategy for bioactive compounds for more effective plant growth regulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. RNA sequencing data associated with this work is available at BioProject accession PRJNA1029024 (manuscript refs. 90,130). Source data are provided with this paper.
References
Verstraeten, I., Schotte, S. & Geelen, D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 5, 495 (2014).
Hartmann, H. T., Kester, D. E., Davis, F. T., Geneve, R. L. & Wilson, S. B. Hartmann & Kester’s Plant Propagation: Principles and Practices (Pearson, 2017).
Poethig, R. S. Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930 (1990).
Pijut, P. M., Woeste, K. E. & Michler, C. H. Promotion of adventitious root formation of difficult-to-root hardwood tree species. Hortic. Rev. 38, 213 (2011).
Hackett, W. P. Juvenility, maturation, and rejuvenation in woody plants. Hortic. Rev. 7, 109–154 (2011).
Wilcox, J. R. & Farmer, R. E. Heritability and C effects in early root growth of eastern cottonwood cuttings. Heredity 23, 239–245 (1968).
Grattapaglia, D., Bertolucci, F. L. & Sederoff, R. R. Genetic mapping of QTLs controlling vegetative propagation in Eucalyptus grandis and E. urophylla using a pseudo-testcross strategy and RAPD markers. Theor. Appl. Genet. 90, 933–947 (1995).
Visser, E. J. W., Cohen, J. D., Barendse, G. W. M., Blom, C. W. P. M. & Voesenek, L. A. C. J. An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiol. 112, 1687–1692 (1996).
Sorin, C. et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17, 1343–1359 (2005).
Bellini, C., Pacurar, D. I. & Perrone, I. Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666 (2014).
Pacurar, D. I., Perrone, I. & Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 151, 83–96 (2014).
Caboni, E. et al. Biochemical aspects of almond microcuttings related to in vitro rooting ability. Biol. Plant. 39, 91–97 (1997).
Lakehal, A. et al. A molecular framework for the control of adventitious rooting by TIR1/AFB2-Aux/IAA-dependent auxin signaling in Arabidopsis. Mol. Plant 12, 1499–1514 (2019).
Bellamine, J., Penel, C., Greppin, H. & Gaspar, T. Confirmation of the role of auxin and calcium in the late phases of adventitious root formation. Plant Growth Regul. 26, 191–194 (1998).
Blažková, A. et al. Auxin metabolism and rooting in young and mature clones of Sequoia sempervirens. Physiol. Plant. 99, 73–80 (1997).
Rasmussen, A., Hosseini, S. A., Hajirezaei, M.-R., Druege, U. & Geelen, D. Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J. Exp. Bot. 66, 1437–1452 (2015).
Abu-Abied, M. et al. Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of Eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. Plant J. 71, 787–799 (2012).
Abarca, D. et al. The GRAS gene family in pine: transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots. BMC Plant Biol. 14, 354 (2014).
Fahn, A. Plant Anatomy (Pergamon, 1990).
Uggla, C., Moritz, T., Sandberg, G. & Sundberg, B. Auxin as a positional signal in pattern formation in plants. Proc. Natl Acad. Sci. USA 93, 9282–9286 (1996).
Tuominen, H., Puech, L., Fink, S. & Sundberg, B. A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 115, 577–585 (1997).
Ballester, A., San-José, M. C., Vidal, N., Fernández-Lorenzo, J. L. & Vieitez, A. M. Anatomical and biochemical events during in vitro rooting of microcuttings from juvenile and mature phases of chestnut. Ann. Bot. 83, 619–629 (1999).
Vidal, N., Arellano, G., San-José, M. C., Vieitez, A. M. & Ballester, A. Developmental stages during the rooting of in-vitro-cultured Quercus robur shoots from material of juvenile and mature origin. Tree Physiol. 23, 1247–1254 (2003).
Legué, V., Rigal, A. & Bhalerao, R. P. Adventitious root formation in tree species: involvement of transcription factors. Physiol. Plant. 151, 192–198 (2014).
Ranjan, A. et al. Molecular basis of differential adventitious rooting competence in poplar genotypes. J. Exp. Bot. 73, 4046–4064 (2022).
Díaz-Sala, C. Direct reprogramming of adult somatic cells toward adventitious root formation in forest tree species: the effect of the juvenile–adult transition. Front. Plant Sci. 5, 310 (2014).
Solé, A. et al. Characterization and expression of a Pinus radiata putative ortholog to the Arabidopsis SHORT-ROOT gene. Tree Physiol. 28, 1629–1639 (2008).
Abu-Abied, M. et al. Gene expression profiling in juvenile and mature cuttings of Eucalyptus grandis reveals the importance of microtubule remodeling during adventitious root formation. BMC Genom. 15, 826 (2014).
de Almeida, M. R. et al. Reference gene selection for quantitative reverse transcription–polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 11, 73 (2010).
de Almeida, M. R. et al. Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in Eucalyptus. Plant Sci. 239, 155–165 (2015).
Ruedell, C. M., de Almeida, M. R. & Fett-Neto, A. G. Concerted transcription of auxin and carbohydrate homeostasis-related genes underlies improved adventitious rooting of microcuttings derived from far-red treated Eucalyptus globulus Labill mother plants. Plant Physiol. Biochem. 97, 11–19 (2015).
Cooper, W. C. Hormones in relation to root formation on stem cuttings. Plant Physiol. 10, 789 (1935).
Oinam, G., Yeung, E., Kurepin, L., Haslam, T. & Lopez-Villalobos, A. Adventitious root formation in ornamental plants: I. General overview and recent successes. Propag. Ornam. Plants 11, 78–90 (2011).
Epstein, E. & Ludwig-Müller, J. Indole-3-butyric acid in plants: occurrence, synthesis, metabolism and transport. Physiol. Plant 88, 382–389 (1993).
Strader, L. C. & Bartel, B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4, 477–486 (2011).
Wiesman, Z., Riov, J. & Epstein, E. Comparison of movement and metabolism of indole‐3‐acetic acid and indole‐3‐butyric acid in mung bean cuttings. Physiol. Plant 74, 556–560 (1988).
Felker, P. & Clark, P. R. Rooting of mesquite (Prosopis) cuttings. J. Range Manag. 34, 466–468 (1981).
Van der Krieken, W. M. et al. in Biology of Root Formation and Development (eds Altman, Q. & Waisel, Y.) 95–104 (Springer, 1997).
Mihaljevic, S. & Salopek-Sondi, B. Alanine conjugate of indole-3-butyric acid improves rooting of highbush blueberries. Plant Soil Environ. 58, 236–241 (2012).
Haissig, B. E. Influence of aryl esters of indole-3-acetic and indole-3-butyric acids on adventitious root primordium initiation and development. Physiol. Plant 47, 29–33 (1979).
Pizarro, A. & Díaz-Sala, C. Cellular dynamics during maturation-related decline of adventitious root formation in forest tree species. Physiol. Plant 165, 73–80 (2019).
Vilasboa, J., da Costa, C. T. & Fett-Neto, A. G. Rooting of eucalypt cuttings as a problem-solving oriented model in plant biology. Prog. Biophys. Mol. Biol. 146, 85–97 (2019).
Quareshy, M., Prusinska, J., Li, J. & Napier, R. A cheminformatics review of auxins as herbicides. J. Exp. Bot. 69, 265–275 (2018).
Eyer, L. et al. 2,4-D and IAA amino acid conjugates show distinct metabolism in Arabidopsis. PLoS ONE 11, e0159269 (2016).
Yang, Y., Hammes, U. Z., Taylor, C. G., Schachtman, D. P. & Nielsen, E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16, 1123–1127 (2006).
Hoyerova, K. et al. Auxin molecular field maps define AUX1 selectivity: many auxin herbicides are not substrates. New Phytol. 217, 1625–1639 (2018).
Calderón Villalobos, L. I. A. et al. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 (2012).
Lee, S. et al. Defining binding efficiency and specificity of auxins for SCFTIR1/AFB-Aux/IAA co-receptor complex formation. ACS Chem. Biol. 9, 673–682 (2014).
Vain, T. et al. Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc. Natl Acad. Sci. USA 116, 6463–6472 (2019).
Pufky, J., Qiu, Y., Rao, M. V., Hurban, P. & Jones, A. M. The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct. Integr. Genom 3, 135–143 (2003).
Delargy, J. A. & Wright, C. E. Root formation in cuttings of apple in relation to auxin application and to etiolation. New Phytol. 82, 341–347 (1979).
Verstraeten, I., Beeckman, T. & Geelen, D. in Plant Organogenesis (ed Ive De Smet) 159–175 (Springer, 2013).
Grossmann, K. Auxin herbicides: current status of mechanism and mode of action. Pest Manag. Sci. 66, 113–120 (2010).
Ludwig-Müller, J., Vertocnik, A. & Town, C. D. Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot. 56, 2095–2105 (2005).
Blythe, E. K., Sibley, J. L., Tilt, K. M. & Ruter, J. M. Methods of auxin application in cutting propagation: a review of 70 years of scientific discovery and commercial practice. J. Environ. Hortic 25, 166–185 (2007).
Riov, J. et al. Improved method for vegetative propagation of mature Pinus halepensis and its hybrids by cuttings. Isr. J. Plant Sci. 67, 5–15 (2020).
Eliasson, L. & Areblad, K. Auxin effects on rooting in pea cuttings. Physiol. Plant 61, 293–297 (1984).
Wain, R. L., Wightman, F. & Russell, E. J. The growth-regulating activity of certain ω-substituted alkyl carboxylic acids in relation to their β-oxidation within the plant. Proc. R. Soc. Lond. B 142, 525–536 (1954).
Behrens, R. & Howard Morton, L. Some factors influencing activity of 12 phenoxy acids on mesquite root inhibition. Plant Physiol. 38, 165–170 (1963).
Zimmerman, P. W. Several chemical growth substances which cause initiation of roots and other responses in plants. Contrib. Boyce Thompson Inst. 7, 209–229 (1935).
Katekar, G. F. Auxins: on the nature of the receptor site and molecular requirements for auxin activity. Phytochemistry 18, 223–233 (1979).
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005).
Xuan, W., Opdenacker, D., Vanneste, S. & Beeckman, T. in Root Development (eds Ristova, D. & Barbez, E.) 177–190 (Springer, 2018).
Ung, K. L. et al. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 609, 605–610 (2022).
Bainbridge, K. et al. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 22, 810–823 (2008).
Casanova-Sáez, R. et al. Inactivation of the entire Arabidopsis group II GH3s confers tolerance to salinity and water deficit. New Phytol. 235, 263–275 (2022).
Hayashi, K. et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 12, 1–11 (2021).
Bartel, B. & Fink, G. R. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748 (1995).
Davies, R. T., Goetz, D. H., Lasswell, J., Anderson, M. N. & Bartel, B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376 (1999).
LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P. T. & Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 (2002).
Campanella, J. J., Olajide, A. F., Magnus, V. & Ludwig-Muller, J. A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol. 135, 2230–2240 (2004).
Rampey, R. A. et al. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988 (2004).
Bitto, E. et al. X‐ray structure of ILL2, an auxin‐conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74, 61–71 (2009).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Valentini, R., Mugnozza, G. S., Giordano, E. & Kuzminsky, E. Influence of cold hardening on water relations of three Eucalyptus species. Tree Physiol. 6, 1–10 (1990).
Eliyahu, A. et al. Vegetative propagation of elite Eucalyptus clones as food source for honeybees (Apis mellifera); adventitious roots versus callus formation. Isr. J. Plant Sci. 67, 83–97 (2020).
Robinson, T. L. et al. Performance of Cornell-Geneva rootstocks across North America in multi-location NC-140 rootstock trials. In Proc. 1st International Symposium on Rootstocks for Deciduous Fruit Tree Species 241–245 (ISHS, 2004).
Marini, R. P. et al. Performance of ‘golden delicious’ apple on 23 rootstocks at 12 locations: a five-year summary of the 2003 nc-140 dwarf rootstock trial. J. Am. Pomol. Soc. 63, 115 (2009).
Marini, R. P. et al. Performance of ‘golden delicious’ apple on 23 rootstocks at eight locations: a ten-year summary of the 2003 NC-140 dwarf rootstock trial. J. Am. Pomol. Soc. 68, 54–68 (2014).
Tzeela, P. et al. Comparing adventitious root-formation and graft-unification abilities in clones of Argania spinosa. Front. Plant Sci. 13, 1002703 (2022).
Ruberv, P. H. & Sheldrake, A. R. Effect of pH and surface charge on cell uptake of auxin. Nat. New Biol. 244, 285–288 (1973).
Sanchez Carranza, A. P. et al. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 6, 24212 (2016).
Staswick, P. E. et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627 (2005).
Hagen, G. & Guilfoyle, T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385 (2002).
Wabnik, K., Govaerts, W., Friml, J. & Kleine-Vehn, J. Feedback models for polarized auxin transport: an emerging trend. Mol. Biosyst. 7, 2352–2359 (2011).
Hudson, J. P. Propagation of plants by root cuttings: II. Seasonal fluctuation of capacity to regenerate from roots. J. Hortic. Sci. 30, 242–251 (1955).
Ohkouchi, T. & Tsuji, K. Basic technology and recent trends in agricultural formulation and application technology. J. Pest Sci. 47, 155–171 (2022).
Skůpa, P., Opatrný, Z. & Petrášek, J. in Applied Plant Cell Biology (eds Nick, P. & Opatrny, Z.) 69–102 (Springer, 2014).
Kessel, A. & Ben-Tal, N. Free energy determinants of peptide association with lipid bilayers. Curr. Top. Membr. 52, 205–253 (2002).
Ridoutt, B. G., Pharis, R. P. & Sands, R. Identification and quantification of cambial region hormones of Eucalyptus globulus. Plant Cell Physiol. 36, 1143–1147 (1995).
Foucart, C. et al. Transcript profiling of a xylem vs phloem cDNA subtractive library identifies new genes expressed during xylogenesis in Eucalyptus. New Phytol. 170, 739–752 (2006).
FastX-Toolkit. Hannon Lab (2012) http://hannonlab.cshl.edu/fastx_toolkit/index.html
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, 1–13 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Moura, J. C. M. S. et al. Validation of reference genes from Eucalyptus spp. under different stress conditions. BMC Res. Notes 5, 634 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Bennett, M. J. et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950 (1996).
Moreno-Risueno, M. A. et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010).
Heisler, M. G. et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P. T. & Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 (2002).
Sela, I., Ashkenazy, H., Katoh, K. & Pupko, T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14 (2015).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Yu, G., Smith, D. K., Zhu, H. & Guan, Y. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Wang, L.-G. et al. Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 37, 599–603 (2020).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019).
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D. G. & Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693 (2013).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6, e16765 (2011).
Grützner, R. et al. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2, 100135 (2021).
Karimi, M., Inzé, D. & Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Zhang, X., Henriques, R., Lin, S.-S., Niu, Q.-W. & Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006).
Prigge, M. J., Lavy, M., Ashton, N. W. & Estelle, M. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 20, 1907–1912 (2010).
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Pierre-Jerome, E., Jang, S. S., Havens, K. A., Nemhauser, J. L. & Klavins, E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl Acad. Sci. USA 111, 9407–9412 (2014).
Prusinska, J. et al. The differential binding and biological efficacy of auxin herbicides. Pest Manag. Sci. 79, 305–1315 (2022).
Fastner, A., Absmanner, B. & Hammes, U. Z. in Plant Hormones (eds Kleine-Vehn, J. & Sauer, M.) 259–270 (Springer, 2017).
Frank, O., Kreissl, J. K., Daschner, A. & Hofmann, T. Accurate determination of reference materials and natural isolates by means of quantitative 1H NMR spectroscopy. J. Agric. Food Chem. 62, 2506–2515 (2014).
Friesner, R. A. et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 49, 6177–6196 (2006).
Halgren, T. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004).
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
Madhavi Sastry, G., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013).
Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Greenwood, J. R., Calkins, D., Sullivan, A. P. & Shelley, J. C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 24, 591–604 (2010).
ChemAxon Chemicalize. ChemAxon (2023) https://disco.chemaxon.com/calculators/demo/plugins/logd/
Pan, X., Wang, H., Li, C., Zhang, J. Z. H. & Ji, C. MolGpka: a web server for small molecule pKa prediction using a graph-convolutional neural network. J. Chem. Inf. Model. 61, 3159–3165 (2021).
Roth, O. et al. Slow release of a synthetic auxin induces formation of adventitious roots in recalcitrant woody plants. Cambium-enriched samples of Eucalyptus grandis mature cuttings BioProject https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1029024 (2023).
Acknowledgements
We thank the undergraduate students A. Verblun and G. Yehezkely in the laboratory of R.W. for supporting experiments. We also thank E. Shani for providing DR5:Luciferase, DR5:Venus and aux1–7 Arabidopsis lines; M. Bennett and R. Swarup for providing the aux/lax quadruple mutant; M. Estelle for providing Y2H vectors; K. Ljung for providing the gh3 octuple mutant and B. Bartel for providing the ilr1-1 ill2-1 iar3-2 triple mutant and vectors expressing GST-recombinant version of ILR1, ILL2 and IAR3. We also thank I. Mayrose and K. Halabi for their assistance in constructing ILR1/ILLs phylogenetics. This work was supported by funding from the Israel Science Foundation (grant number 1057/21 to R.W.), the Chief Scientist of the Ministry of Agriculture and Rural Development, Israel (grant numbers 20-10-0067, 13-37-0005 and 20-01-0270 to R.W. and E.S.), the United States–Israel Binational Agricultural Research and Development Fund (BARD, grant number IS-5195-19R to R.W, E.S and C.J. Staiger (Purdue University, IN)), the Yuri Milner 70@70 Fellowship (to O.R.), the Deutsche Forschungsgemeinschaft (SFB024 TPB12 to C.D and SFB924 TPA08 and HA3468/6-3 to U.Z.H) and the Tel Aviv University Center for AI and Data Science (to N.B.-T).
Author information
Authors and Affiliations
Contributions
J.R., E.S. and R.W. conceived of the project and E.S. and R.W. supervised the project. O.R. designed and ran experiments and analyzed data. S.Y., O.S., A.E. and P.T. ran rooting experiments of cuttings from the different trees. I.V. synthesized and characterized compounds. A.E. and V.D. performed RNA sequencing and qPCR experiments. F.S. ran mass-spectrometry analyses of auxins and conjugates and analyzed data under the supervision of M.C.-W. A.K. designed and ran molecular simulation experiments with input from O.R. under the supervision of N.B.-T. A.F.-D. performed bioinformatics analysis of the RNA sequencing data. R.N. designed and performed SPR measurements. D.P.J. performed oocyte uptake assay under the supervision of U.Z.H. U.Z.H. performed SSM assays. V.P. performed mass spectrometry analyses on oocyte extracts and membranes under supervision of C.D. K.L.U. performed PIN purification under supervision of B.P.P. A.C. performed and analyzed binding assays of auxin receptors under the supervision of E.K. E.S., R.W. and O.R. wrote the manuscript with inputs from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Danny Geelan, Catherine Bellini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–26, Tables 1–3 and 1H- and 13C-NMR and mass-spectrometry data.
Supplementary Data
Source data for supplementary figures.
Source data
Source Data Figs. 1–6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Roth, O., Yechezkel, S., Serero, O. et al. Slow release of a synthetic auxin induces formation of adventitious roots in recalcitrant woody plants. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-023-02065-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-02065-3
This article is cited by
-
Horticultural potential of chemical biology to improve adventitious rooting
Horticulture Advances (2024)