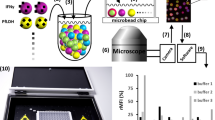
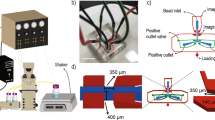
Abstract
Aptamers, commonly referred to as chemical antibodies, are used in a wide range of applications including drug delivery and biosensing. However, the process of aptamer selection poses a substantial challenge, as it requires numerous cycles of enrichment and involves issues with nonspecific binding. We present a simple, fast instrument-free method for aptamer enrichment and selection based on a diffusion-binding process in a three-dimensional non-fouling porous hydrogel with immobilized target proteins. Low-affinity aptamer candidates can be rapidly released from the hydrogel, whereas high-affinity candidates are restricted due to their strong binding to the immobilized protein targets. Consequently, a one-step enriched aptamer pool can strongly bind the protein targets. This enrichment is consistent across five proteins with isoelectric points in varying ranges. With thrombin as a representative model, the anti-thrombin aptamer identified from an enriched aptamer pool has been found to have a binding affinity that is comparable to those identified over ten cycles of selection using traditional methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data supporting this new method are available within the article and its Supplementary Information. Parameters for simulation are provided in Supplementary Table 1. Sequences of ssDNA library, PCR primers and aptamer candidates are included in Supplementary Tables 2–5. All uncropped gel electrophoresis data are available in Source Data and Supplementary Information. Source data are provided with this paper.
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 1742–1749 (2017).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Benenson, Y., Adar, R., Paz-Elizur, T., Livneh, Z. & Shapiro, E. DNA molecule provides a computing machine with both data and fuel. Proc. Natl Acad. Sci. USA 100, 2191–2196 (2003).
Li, Y. et al. Controlled assembly of dendrimer-like DNA. Nat. Mater. 3, 38–42 (2004).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Tasset, D., Kubik, M. F. & Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 272, 688–698 (1997).
Boshtam, M., Asgary, S., Kouhpayeh, S., Shariati, L. & Khanahmad, H. Aptamers against pro- and anti-inflammatory cytokines: a review. Inflammation 40, 340–349 (2017).
Famulok, M. & Mayer, G. Aptamers and SELEX in chemistry and biology. Chem. Biol. 21, 1055–1058 (2014).
Rusconi, C. P. et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419, 90–94 (2002).
Farokhzad, O. C. et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl Acad. Sci. USA 103, 6315–6320 (2006).
Liu, J. & Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. 45, 90–94 (2006).
Zhao, W. et al. Cell-surface sensors for real-time probing of cellular environments. Nat. Nanotechnol. 6, 524–531 (2011).
McNamara, J. A. et al. Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006).
Gotrik, M. R., Feagin, T. A., Csordas, A. T., Nakamoto, M. & Soh, H. T. Advancements in aptamer discovery technologies. Acc. Chem. Res. 49, 1903–1910 (2016).
Vant-Hull, B. P., Payano-Baez, A., Davis, R. H. & Gold, L. The mathematics of SELEX against complex targets. J. Mol. Biol. 278, 579–597 (1998).
Djordjevic, M. SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol. Eng 24, 179–189 (2007).
Mendonsa, S. D. & Bowser, M. T. In vitro selection of aptamers with affinity for neuropeptide y using capillary electrophoresis. J. Am. Chem. Soc. 127, 9382–9383 (2005).
Hoon, S., Zhou, B., Janda, K. D., Brenner, S. & Scolnick, J. Aptamer selection by high-throughput sequencing and informatic analysis. BioTechniques 51, 413–416 (2011).
Lou, X. et al. Micromagnetic selection of aptamers in microfluidic channels. PNAS 106, 2989–2994 (2009).
Kim, J. et al. Integrated microfluidic isolation of aptamers using electrophoretic oligonucleotide manipulation. Sci. Rep. 6, 26139 (2016).
Wilson, B. & Soh, H. T. Re-evaluating the conventional wisdom about binding assays. Trends Biochem. Sci. 45, 639–649 (2020).
Sun, Y., Gombotz, W. R. & Hoffman, A. Synthesis and characterization of non-fouling polymer surfaces: I. Radiation grafting of hydroxyethyl methacrylate and polyethylene glycol methacrylate onto silastic film. J. Bioact. Compat. Polym. 1, 316–334 (1986).
Ma, H., Hyun, J., Stiller, P. & Chilkoti, A. ‘Non-fouling’ oligo(ethylene glycol)-functionalized polymer brushes synthesized by surface-initiated atom transfer radical polymerization. Adv. Mater. 16, 338–341 (2004).
Sanchez-Cano, C. & Carril, M. Recent developments in the design of non-biofouling coatings for nanoparticles and surfaces. Int. J. Mol. Sci. 21, 1007 (2020).
MacDonald, J., Houghton, P., Xiang, D., Duan, W. & Shigdar, S. Truncation and mutation of a transferrin receptor aptamer enhances binding affinity. Nucleic Acid Ther. 26, 348–354 (2016).
Le, T. T., Chumphukam, O. & Cass, A. Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Adv. 4, 47227–47233 (2014).
Bock, L. C., Griffin, L. L., Latham, J., Vermaas, E. & Toole, J. M. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992).
Tokmakov, A. A., Kurotani, A. & Sato, K. Protein pI and intracellular localization. Front. Mol. Biosci. 8, 775736 (2021).
Ma, X., Balasubramanian, G. & Shrotriya, P. Electrical stimulus controlled binding/unbinding of human thrombin-aptamer complex. Sci. Rep. 6, 37449 (2016).
Jiang, Z., You, L., Dou, W., Sun, T. & Xu, P. Effects of an electric field on the conformational transition of the protein: a molecular dynamics simulation study. Polymers 11, 282 (2019).
Thormann, W., Zhang, C., Caslavska, J., Gebauer, P. & Mosher, R. A. Modeling of the impact of ionic strength on the electroosmotic flow in capillary electrophoresis with uniform and discontinuous buffer systems. Anal. Chem. 70, 549–562 (1998).
Acinas, S. G., Sarma-Rupavtarm, R. B., Klepac-Ceraj, V. & Polz, M. F. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71, 8966–8969 (2005).
Takahashi, M. et al. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci. Rep. 6, 33697 (2016).
Cho, M. et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. PNAS 107, 15373–15378 (2010).
Kramer, S. T., Gruenke, P. R., Alam, K. K., Xu, D. & Burke, D. H. FASTAptameR 2.0: a web tool for combinatorial sequence selections. Mol. Ther. Nucleic Acids 29, 862–870 (2022).
Acknowledgements
This work was partly supported by the National Institutes of Health (grants R01HL122311 and R01AR073364).
Author information
Authors and Affiliations
Contributions
N.K.S. designed experiments, did experiments, analyzed data and wrote the manuscript. Yixun Wang studied the simulation of aptamer transport, did experiments, analyzed data and wrote the manuscript. C.W., B.D., X.W. and K.L. performed experiments and analyzed data. Yong Wang coined the concept, designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
N.K.S. and Yong Wang filed a patent application (US patent 63/428,466) based on the method presented in this work.
Peer review
Peer review information
Nature Biotechnology thanks Honggang Cui, John Rossi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Tables 1–6.
Supplementary Video 1
Supporting video 1.
Source data
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 4
Unprocessed gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, N.K., Wang, Y., Wen, C. et al. High-affinity one-step aptamer selection using a non-fouling porous hydrogel. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-023-01973-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-01973-8