Abstract
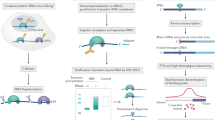
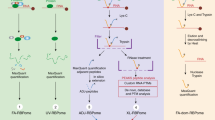
RNA–protein interaction networks govern many biological processes but are difficult to examine comprehensively. We devised ribonucleoprotein networks analyzed by mutational profiling (RNP-MaP), a live-cell chemical probing strategy that maps cooperative interactions among multiple proteins bound to single RNA molecules at nucleotide resolution. RNP-MaP uses a hetero-bifunctional crosslinker to freeze interacting proteins in place on RNA and then maps multiple bound proteins on single RNA strands by read-through reverse transcription and DNA sequencing. RNP-MaP revealed that RNase P and RMRP, two sequence-divergent but structurally related non-coding RNAs, share RNP networks and that network hubs define functional sites in these RNAs. RNP-MaP also identified protein interaction networks conserved between mouse and human XIST long non-coding RNAs and defined protein communities whose binding sites colocalize and form networks in functional regions of XIST. RNP-MaP enables discovery and efficient validation of functional protein interaction networks on long RNAs in living cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw and processed sequencing data sets analyzed in this report will be made available upon reasonable request and have been deposited in the Gene Expression Omnibus database (GSE152483).
Code availability
ShapeMapper2, ΔSHAPE, SuperFold and RingMapper software used for analysis are available at http://weeks.chem.unc.edu/software.html and https://github.com/Weeks-UNC. MEME, VARNA, PyMol and Gephi are all third-party, open-source software.
References
Gehring, N. H., Wahle, E. & Fischer, U. Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem. Sci. 42, 369–382 (2017).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012).
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013).
Pomeranz Krummel, D. A., Oubridge, C., Leung, A. K. W., Li, J. & Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 resolution. Nature 458, 475–480 (2009).
Kondo, Y., Oubridge, C., van Roon, A. M. M. & Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4, 1–19 (2015).
Wu, J. et al. Cryo-EM structure of the human ribonuclease P holoenzyme. Cell 175, 1393–1404.e11 (2018).
Ule, J., Hwang, H. W. & Darnell, R. B. The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol. 10, a032243 (2018).
Garzia, A., Meyer, C., Morozov, P., Sajek, M. & Tuschl, T. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods 118–119, 24–40 (2017).
Wheeler, E. C., Van Nostrand, E. L. & Yeo, G. W. Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip. Rev. RNA 9, e1436 (2018).
Freeberg, M. A. et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 14, R13 (2013).
Schueler, M. et al. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol. 15, R15 (2014).
Ramanathan, M., Porter, D. F. & Khavari, P. A. Methods to study RNA–protein interactions. Nat. Methods 16, 225–234 (2019).
Mädler, S., Bich, C., Touboul, D. & Zenobi, R. Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J. Mass Spectrom. 44, 694–706 (2009).
Das, J. Aliphatic diazirines as photoaffinity probes for proteins: recent developments. Chem. Rev. 111, 4405–4417 (2011).
Krüger, D. M., Neubacher, S. & Grossmann, T. N. Protein–RNA interactions: structural characteristics and hotspot amino acids. RNA 24, 1457–1465 (2018).
Smola, M. J., Rice, G. M., Busan, S., Siegfried, N. A. & Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669 (2015).
Smola, M. J. et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl Acad. Sci. USA 113, 10322–10327 (2016).
Yang, T., Li, X. M., Bao, X., Fung, Y. M. E. & Li, X. D. Photo-lysine captures proteins that bind lysine post-translational modifications. Nat. Chem. Biol. 12, 70–72 (2016).
Kühn-Hölsken, E. et al. Mapping the binding site of snurportin 1 on native u1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res. 38, 5581–5593 (2010).
Mustoe, A. M., Lama, N. N., Irving, P. S., Olson, S. W. & Weeks, K. M. RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl Acad. Sci. USA 116, 24574–24582 (2019).
So, B. R. et al. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 23, 225–230 (2016).
Will, C. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24, 4614–4623 (1996).
Esakova, O. & Krasilnikov, A. S. Of proteins and RNA: the RNase P/MRP family. RNA 16, 1725–1747 (2010).
Perederina, A., Berezin, I. & Krasilnikov, A. S. In vitro reconstitution and analysis of eukaryotic RNase P RNPs. Nucleic Acids Res. 46, 6857–6868 (2018).
Sahakyan, A., Yang, Y. & Plath, K. The role of Xist in X-chromosome dosage compensation. Trends Cell Biol. 28, 999–1013 (2018).
Engreitz, J. M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013).
Wutz, A., Rasmussen, T. P. & Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174 (2002).
Colognori, D., Sunwoo, H., Kriz, A. J., Wang, C. Y. & Lee, J. T. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell 74, 101–117 (2019).
Ridings-Figueroa, R. et al. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31, 876–888 (2017).
Sunwoo, H., Colognori, D., Froberg, J. E., Jeon, Y. & Lee, J. T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl Acad. Sci. USA 114, 10654–10659 (2017).
Lee, H. J. et al. En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements. Nucleic Acids Res. 47, 3875–3887 (2019).
Nesterova, T. B. et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10, 3129 (2019).
Brockdorff, N. Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Noncoding RNA 4, 28 (2018).
Davis, C. A. et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801 (2018).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Moindrot, B. et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572 (2015).
Ciaudo, C. et al. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2, e94 (2006).
Sakaguchi, T. et al. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev. Cell 39, 11–12 (2016).
Patil, D. P. et al. M6 A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Xiao, R. et al. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121 (2019).
Yap, K. et al. A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol. Cell 72, 525–540 (2018).
Rayman, J. B., Karl, K. A. & Kandel, E. R. TIA-1 self-multimerization, phase separation, and recruitment into stress granules are dynamically regulated by Zn 2. Cell Rep. 22, 59–71 (2018).
Gallego-Iradi, M. C. et al. N-terminal sequences in matrin 3 mediate phase separation into droplet-like structures that recruit TDP43 variants lacking RNA binding elements. Lab. Investig. 99, 1030–1040 (2019).
Attig, J. et al. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 174, 1067–1081 (2018).
Long, J. C. & Caceres, J. F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27 (2009).
De, I. et al. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol. 22, 138–144 (2015).
Rigo, F. et al. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat. Chem. Biol. 8, 555–561 (2012).
Sugimoto, N. et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res. 43, 5898–5911 (2015).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Dominguez, D. et al. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell 70, 854–867 (2018).
Xue, Y. et al. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36, 996–1006 (2009).
Uemura, Y. et al. Matrin3 binds directly to intronic pyrimidine-rich sequences and controls alternative splicing. Genes Cells 22, 785–798 (2017).
Meyer, C. et al. The TIA1 RNA-binding protein family regulates EIF2AK2-mediated stress response and cell cycle progression. Mol. Cell 69, 622–635 (2018).
Cerase, A. et al. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 26, 331–334 (2019).
Pandya-Jones, A. et al. A protein assembly mediates Xist localization and silencing. Nature https://doi.org/10.1038/s41586-020-2703-0 (2020).
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R. & Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548 (2018).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks visualization and exploration of large graphs. Int. AAAI Conf. Weblogs Soc. Media https://doi.org/10.13140/2.1.1341.1520 (2009).
Blondel, V. D., Guillaume, J. L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. P10008 (2008).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proc. Second International Conference on Intelligent Systems for Molecular Biololgy 2, 28–36 (1994).
Busan, S., Weidmann, C. A., Sengupta, A. & Weeks, K. M. Guidelines for SHAPE reagent choice and detection strategy for RNA structure probing studies. Biochemistry 58, 2655–2664 (2019).
Sengupta, A., Rice, G. M. & Weeks, K. M. Single-molecule correlated chemical probing reveals large-scale structural communication in the ribosome and the mechanism of the antibiotic spectinomycin in living cells. PLoS Biol. 17, e3000393 (2019).
Busan, S. & Weeks, K. M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018).
Reuter, J. S. & Mathews, D. H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129 (2010).
Smola, M. J., Calabrese, J. M. & Weeks, K. M. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54, 6867–6875 (2015).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2011).
DeLano, W. Pymol: an open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography 40, 82–92 (2002).
Darty, K., Denise, A. & Ponty, Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Acknowledgements
The work was supported by grants from the National Science Foundation (MCB-1121024) and the National Institutes of Health (R35 GM122532) to K.M.W. C.A.W. is a postdoctoral fellow at the American Cancer Society (ACS 130845-RSG-17-114-01-RMC). J.M.C. was supported by National Institutes of Health grant R01 GM121806. Xist and XIST antisense probes were provided by the M. Guttman laboratory (CalTech), and we thank M. Blanco (CalTech) for his initial support in their application. XIST eCLIP data from published works were provided by the G.W. Yeo laboratory (UCSD), and we thank G.W. Yeo (UCSD), M. Corley (UCSD) and D. Sprague (UNC) for support in formatting these data for integration into this work and for helpful comments on the project.
Author information
Authors and Affiliations
Contributions
C.A.W. and P.B.J. conducted experiments. C.A.W., A.M.M. and K.M.W. analyzed data. C.A.W., J.M.C. and K.M.W. designed and interpreted experiments. The manuscript was written by C.A.W. and K.M.W. with input from all authors.
Corresponding author
Ethics declarations
Competing interests
A.M.M. is an advisor to and K.M.W. is an advisor to and holds equity in Ribometrix, to which mutational profiling technologies have been licensed.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and legends for Supplementary Data 1 and 2 and Supplementary Tables 1–3.
Supplementary Data 1
RNaseP-RMRP structural alignment
Supplementary Data 2
eCLIP sites used for analysis
Supplementary Table 1
MI linking eCLIP
Supplementary Table 2
Significantly excluded eCLIP pairs
Supplementary Table 3
Reporter and primer sequences
Rights and permissions
About this article
Cite this article
Weidmann, C.A., Mustoe, A.M., Jariwala, P.B. et al. Analysis of RNA–protein networks with RNP-MaP defines functional hubs on RNA. Nat Biotechnol 39, 347–356 (2021). https://doi.org/10.1038/s41587-020-0709-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0709-7