Abstract
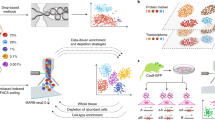
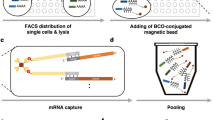
We describe a universal sample multiplexing method for single-cell RNA sequencing in which fixed cells are chemically labeled by attaching identifying DNA oligonucleotides to cellular proteins. Analysis of a 96-plex perturbation experiment revealed changes in cell population structure and transcriptional states that cannot be discerned from bulk measurements, establishing an efficient method for surveying cell populations from large experiments or clinical samples with the depth and resolution of single-cell RNA sequencing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Sequencing data from these experiments can be obtained from CaltechDATA at https://doi.org/10.22002/D1.1311.
Code availability
Code and tutorials for the kITE demultiplexing workflow can be found at https://www.kallistobus.tools/kite_tutorial.html. Python notebooks used to process data and generate figures are available on GitHub at https://github.com/pachterlab/GPCTP_2019. The same GitHub repository also contains a fully reproducible reanalysis using ‘kallisto | bustools’ transcript alignments and a Google Colab notebook.
References
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Han, X. et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17 (2018).
Cao, J et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Stoeckius, M. et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
McGinnis, C. S. et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626 (2019).
Guo, C. et al. CellTag indexing: genetic barcode-based sample multiplexing for single-cell genomics. Genome Biol. 20, 90 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Suryawanshi, H. et al. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Bond, A. M., Ming, G. & Song, H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385–395 (2015).
Imayoshi, I. et al. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208 (2013).
Hitoshi, S. et al. Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat. Neurosci. 14, 957–964 (2011).
Yuzwa, S. A. et al. Developmental emergence of adult neural stem cells as revealed by single-cell transcriptional profiling. Cell Rep. 21, 3970–3986 (2017).
Janes, K. A. A Systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310, 1646–1653 (2005).
Nelander, S. et al. Models from experiments: combinatorial drug perturbations of cancer cells. Mol. Syst. Biol. 4, 216 (2008).
Sims, D. et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 12, R104 (2011).
Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Hsiao, S. C. et al. Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir 25, 6985–6991 (2009).
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Melsted, P., Ntranos, V. & Pachter, L. The barcode, UMI, set format and BUStools. Bioinformatics 35(21), 4472–4473 (2019).
Melsted, P. et al. Modular and efficient pre-processing of single-cell RNA-seq. Preprint at bioRxiv https://doi.org/10.1101/673285 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Acknowledgements
We thank Z. Gartner and C. McGinnis for helpful feedback regarding the ClickTag protocol and V. Svensson for suggestions regarding analysis of multiplexed datasets. Thanks to P. Melsted and S. Booeshaghi for developing the ‘kallisto | bustools’ functions used in the preprocessing workflow and to P. Rivaud for assistance with 10x data processing. Additional support was provided by the the Caltech Bioinformatics Resource Center and the Single Cell Profiling and Engineering Center (SPEC) in the Beckman Institute at Caltech.
Author information
Authors and Affiliations
Contributions
J.G. conceived and developed the ClickTag multiplexing strategy. J.G., J.H.P. and S.C. designed the scRNA-seq experiments and J.G. and J.H.P. performed the experiments. J.H.P. performed all tissue culture operations and J.G. developed the kITE demultiplexing workflow and analyzed the scRNA-seq data. J.G., J.H.P., S.C, M.T. and L.P. contributed to the interpretation of the results and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.G., L.P., S.C. and J.H.P. are listed as co-inventors on a patent application related to this work (US patent application 16/296,075).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Materials
Supplementary Figs. 1–19 and Supplementary Tables 1–2.
Supplementary Table 3
Ninety-six-sample multiplex Louvain marker genes. The top 100 marker genes from each cluster for n=21,223 cells in the 96-sample multiplexed experiment. Clusters used for differential expression were determined by Louvain community detection with resolution = 2.2. Differentially expressed genes were identified with the Wilcoxon test and the ScanPy ‘rank_genes_groups’ function.
Supplementary Table 4
Primer sequences used in this study.
Rights and permissions
About this article
Cite this article
Gehring, J., Hwee Park, J., Chen, S. et al. Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat Biotechnol 38, 35–38 (2020). https://doi.org/10.1038/s41587-019-0372-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0372-z
This article is cited by
-
deMULTIplex2: robust sample demultiplexing for scRNA-seq
Genome Biology (2024)
-
DELVE: feature selection for preserving biological trajectories in single-cell data
Nature Communications (2024)
-
scPerturb: harmonized single-cell perturbation data
Nature Methods (2024)
-
High-capacity sample multiplexing for single cell chromatin accessibility profiling
BMC Genomics (2023)
-
FIPRESCI: droplet microfluidics based combinatorial indexing for massive-scale 5′-end single-cell RNA sequencing
Genome Biology (2023)