Abstract
The intestinal immune system is highly adapted to maintaining tolerance to the commensal microbiota and self-antigens while defending against invading pathogens1,2. Recognizing how the diverse network of local cells establish homeostasis and maintains it in the complex immune environment of the gut is critical to understanding how tolerance can be re-established following dysfunction, such as in inflammatory disorders. Although cell and molecular interactions that control T regulatory (Treg) cell development and function have been identified3,4, less is known about the cellular neighbourhoods and spatial compartmentalization that shapes microorganism-reactive Treg cell function. Here we used in vivo live imaging, photo-activation-guided single-cell RNA sequencing5,6,7 and spatial transcriptomics to follow the natural history of T cells that are reactive towards Helicobacter hepaticus through space and time in the settings of tolerance and inflammation. Although antigen stimulation can occur anywhere in the tissue, the lamina propria—but not embedded lymphoid aggregates—is the key microniche that supports effector Treg (eTreg) cell function. eTreg cells are stable once their niche is established; however, unleashing inflammation breaks down compartmentalization, leading to dominance of CD103+SIRPα+ dendritic cells in the lamina propria. We identify and validate the putative tolerogenic interaction between CD206+ macrophages and eTreg cells in the lamina propria and identify receptor–ligand pairs that are likely to govern the interaction. Our results reveal a spatial mechanism of tolerance in the lamina propria and demonstrate how knowledge of local interactions may contribute to the next generation of tolerance-inducing therapies.
Similar content being viewed by others
Main
The intestinal immune system interacts with large numbers of diverse microorganisms. This adaptive interplay between host and microorganism provides a window into how immune tolerance is established and maintained in a complex environment. The pathobiont Helicobacter hepaticus (Hh) establishes lifelong infection in the caecum of wild-type mice. A key host adaptation to ensure immune homeostasis in the face of chronic infection is the production of IL-10 by Treg cells8,9,10. Colonization of previously uninfected mice is used as a model system to study how tolerance is established. Maladaptation of this response results in colitis, and similar processes underlie very early onset inflammatory bowel disease, with deficiencies in the IL-10 pathway as a major cause11. The mesenteric lymph nodes (MLN) have been shown to be a key site of Treg cell induction12,13,14, but the key anatomical location for induction and maintenance of eTreg cell suppressor function is yet to be identified. We used T cell receptor (TCR) transgenic HH7-2tg cells15 (TCRHh) as sentinels in Hh-colonized hosts to follow the natural history of antigen-specific T cells experiencing key interactions and gaining and sustaining Treg cell effector functions in the tissue microenvironment.
eTreg cells function in intestinal tissue
To map where adaptive responses to Hh occur, we characterized the intestinal lymphoid and non-lymphoid tissue compartments of wild-type mice. Lymphoid tissue comprised secondary lymphoid organs, including MLN, the caecal patch (CP) and distal colon organized lymphoid structures (OLS) (Fig. 1a). Small lymphoid aggregates (LAs) are present in the caecum and proximal colon. In line with previous studies16,17, LAs do not have organized T cell and B cell zones and contain a spectrum of tissue organization from cell aggregates to cryptopatches and isolated lymphoid follicles (Extended Data Fig. 1a–c). Previous tracking of naive TCRHh T cells in vivo showed that they differentiated to RORγt+FOXP3+ Treg cells in the colon, but that work did not exclude OLS from the analysis15. We sought to understand the natural history of TCRHh T cells in the context of the lymphoid and non-lymphoid compartments. Here we refer to ‘tissue’ as the compartment comprising lamina propria (LP) and LAs; we compare this with the secondary lymphoid organs: MLN, CP and OLS.
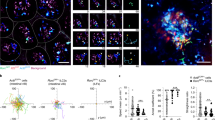
a, Diagram of tissue and lymphoid structures in the mouse gut, with representative images of CP, organized lymphoid structures and LAs in CD2DsRed mouse. Scale bars, 1,000 μm. b, Schematic of transfer of naive TCRHh T cells into Hh-colonized hosts. c, Representative fluorescence-activated cell sorting (FACS) plots of TCRHh T cells at the indicated sites 14 days after transfer of naive TCRHh T cells. d, Frequency of TCRHh cells among total CD4 T cells at indicated timepoints after transfer of naive TCRHh T cells. e, Frequency of Ki-67+ TCRHh cells at the indicated sites 14 days after transfer of naive TCRHh T cells. f, Representative FACS plots (left) and frequency of Treg cells of TCRHh (right) at 14 days after transfer of naive TCRHh T cells. g, Representative histogram overlay (left) and GFP MFI of TCRHhNur77GFP T cells (right) 11 days after transfer of naive TCRHh T cells. h, Representative FACS plots (left) and frequency of Il10+ cells among TCRHh T cells (right) 11 days after transfer of naive TCRHh T cells. i, Representative images of TCRHhNur77GFP (left and centre left) and TCRHhIl10GFP (centre right and right) T cells in the LA and LP. TCRHhNur77GFP and Il10-GFP-positive cells are indicated by white arrowheads. Scale bars, 50 μm. j, Frequency of Nur77GFP (left) and Il10-GFP-positive (right) cells among TCRHh T cells in the LA and LP. a, Representative images from two individual mice over two independent imaging experiments. c–e, Four individual mice representative of two independent experiments. f, Seven individual mice representative of two independent experiments. g,h, Five individual mice representative of two independent experiments. i,j, Representative images and combined data from six individual mice over two independent imaging experiments. Scale bars, 50 μm. e–h, One-way ANOVA using Tukey’s multiple comparisons test. j, Two-tailed t-test.
Naive TCRHh T cells transferred into Hh-colonized hosts (Fig. 1b and Extended Data Fig. 1d) traffic to the secondary lymphoid organs within 24 h and onwards to the tissue approximately 1 week later (Fig. 1c,d). To pinpoint where important cellular interactions and activation steps take place, we quantified proliferation, Treg cell differentiation, TCR signalling and IL-10 production across compartments. Despite exhibiting early activation in the lymphoid compartments (Extended Data Fig. 2a), at 14 days after transfer, TCRHh T cells in the tissue are more proliferative than their lymphoid counterparts (Fig. 1e). A larger proportion of TCRHh T cells differentiated into FOXP3+ Treg cells (Fig. 1f) in the tissue than in the secondary lymphoid organs. TCRHhRag1−/− control donors and uninfected recipients confirmed the specificity of TCRHh T cells and the dependence on cognate antigen for survival (Extended Data Fig. 2b–f).
As Treg cell suppressor function has been shown to depend on TCRs18, we explored whether ongoing TCR stimulation occurs in our defined microenvironments. We crossed TCRHh mice to Nur77GFP mice19 (TCRHhNur77GFP) and transferred T cells from these mice into Hh-colonized hosts; the proportion of cells expressing GFP and the GFP geometric mean fluorescence intensity (MFI) in the total transferred population—measures of TCR signalling—were highest in the tissue at 11 days after the transfer (Fig. 1g). Transfer of T cells from TCRHh mice crossed to C57BL/6 Foxp3CD2Il10GFP reporter mice20 (TCRHhIl10GFP) into Hh-colonized hosts also revealed the tissue as the site with the highest proportion of Il10+TCRHh T cells at day 11 after transfer (Fig. 1h). We next focused on Treg cells, finding similar TCR signalling in TCRHhNur77GFP Treg cells across tissue and secondary lymphoid organs and the highest frequency of TCRHhIl10GFP cells among Treg cells in the tissue (Extended Data Fig. 3a,b).
To determine whether TCR engagement and IL-10 production were a result of local stimulation, we used the sphingosine-1-phosphate receptor agonist FTY720 to block lymph node egress following TCRHh recruitment into the gut to ensure detection of local GFP induction (Extended Data Fig. 3c–e). Again, TCR signalling and the highest IL-10 production were detected in tissue TCRHh T cells, similar to results in untreated controls (Extended Data Fig. 3f–i). Absolute numbers of tissue TCRHh T cells were similar between FTY720-treated mice and untreated controls, suggesting local activation and proliferation of TCRHh T cells (Extended Data Fig. 3j). These data point to the tissue as a site of sustained Treg cell activation and enhanced effector function in the maintenance of tolerance to the Hh. To test the role of the tissue LP microniche in isolation, we used mice deficient in lymphotoxin-α (Lta−/− mice), which are devoid of MLNs, CP, OLS and LA17,21 (Extended Data Fig. 4a and data not shown). Lta−/− hosts supported TCRHh T cell homing to tissue (Extended Data Fig. 4b–d), but in the absence of lymphoid structures, TCRHh T cells did not differentiate into Treg cells (Extended Data Fig. 4e), indicating a non-redundant role for secondary lymphoid organs in Treg cell differentiation. Similar absolute numbers of TCRHh T cells in Lta−/− hosts retained the ability to produce IL-10 compared with controls, albeit at a lower frequency (Extended Data Fig. 4f–h). To determine whether the IL-10 produced in this reductionist system is critical for homeostasis, we blocked IL-10 signalling using a blocking IL-10 receptor (IL10R) monoclonal antibody, which resulted in inflammation (Extended Data Fig. 4i). This shows that IL-10 has a critical role in establishing homeostasis in the LP of Lta−/− mice and that lymph nodes are not required for the pathogenic response to Hh.
Our results demonstrate that tissue interactions have a previously unrecognized role in TCR stimulation and IL-10 production by T cells that are reactive to microorganisms, including Treg cells. Next, we aimed to determine whether the LP and LA niches had distinct roles in shaping the TCRHh response. We bred TCRHhNur77GFP and TCRHhIl10GFP mice with CD2DsRed mice22, which label T cells (resulting in TCRHhCD2DsRedNur77GFP or TCRHhCD2DsRedIl10GFP mice) and separately transferred naive T cells from these reporter lines into Hh-colonized Foxp3GFP hosts23 for in vivo two-photon live imaging (Extended Data Fig. 4j). Host LAs appear as green clusters owing to the presence of host Treg cells. Transferred TCRHh T cells are labelled with DsRed, but co-express GFP upon TCR stimulation or IL-10 production. Three-dimensional imaging enables the visualization of green and red expression from overlapping cells (Supplementary Video 1).
Within tissue, TCRHhNur77GFP+ T cells were present within LAs (Fig. 1i). TCRHhNur77GFP+ cells were also located within the LP, spatially distant from LAs (Fig. 1i). This even distribution of recently activated T cells (Fig. 1j) suggests that TCR–major histocompatibility complex class II (MHCII) interactions occur between TCRHh T cells and LP-resident antigen-presenting cells (APCs), and/or rapid migration of TCR-stimulated TCRHh T cells out of LAs and into LP after TCR engagement. Indeed, time-lapse videos reveal that TCRHhNur77GFP+ T cells are highly motile (Supplementary Video 2). We next examined which tissue niche supported the highest production of IL-1024. In vivo live imaging of donor TCRHhCD2DsRedIl10GFP T cells in Hh-colonized Foxp3GFP hosts indicated that IL-10 production by TCRHh cells was largely restricted to the LP (Fig. 1i,j). TCRHhIl10+ T cells were actively motile throughout the tissue (Supplementary Video 3), suggesting a far-reaching suppressor function in response to Hh.
Spatially and phenotypically distinct Treg cells
Because TCRHh T cells that have recently experienced TCR stimulation are distributed throughout the LP and LA, whereas production of the Treg cell effector molecule IL-10 is highest in the LP, we examined the cellular interactions and/or molecular cues in the LP that drive TCRHh activation and Treg cell effector functions. We used two-photon photo-activation labelling of cells in T cell niches followed by single-cell RNA sequencing (NICHE-seq) to uncover cellular composition and transcriptional states of tissue microniches6,7. Naive T cells from TCRHhCD2DsRed mice crossed to mice ubiquitously expressing photo-activatable GFP6 (TCRHhCD2DsRedUbPA-GFP) were transferred to Hh-colonized CD2DsRedUbPA-GFP hosts so that both host and donor cells were photo-activatable. DsRed was used to visualize donor cells and to mark T cell zones in host lymphoid tissue. We used two-photon microscopy to convert donor and host cells with photo-activated GFP from four regions: the T cell zones of MLNs and CP, LA and LP (Fig. 2a and Extended Data Fig. 5a,b). For each individual mouse, we photo-activated 3–10 regions per tissue location, dissociated tissues, and pooled each tissue location for sorting and sequencing. Single-cell RNA sequencing (scRNA-seq) of GFP+ cells using 10X, 5′ GEX and V(D)J TCR sequencing (Supplementary Data 1). A model built on previous work15,25,26,27,28,29,30,31 supported annotation of a spectrum of myeloid cells, T cells, innate lymphoid cells (ILCs), B cells, plasma cells, epithelial cells, mesenchymal cells, endothelial cells and granulocytes across all four sites (Extended Data Fig. 5c,d, Supplementary Table 1 and Supplementary Data 2–4). The relative distributions of these major intestinal cell types differ according to region, with a sizeable T cell population present across microniches (Extended Data Fig. 5d). Early memory B cells (eMBCs) were present in the LA and absent in the LP, enabling this population to be used to benchmark further LA analyses (Supplementary Data 4).
a, Schematic describing transfer of TCRHhCD2DsRedUbPA-GFP T cells into six CD2DsRedUbPA-GFP hosts with photo-activation, cell sorting and scRNA-seq. DC, dendritic cell; lymph, lymphatic; Mac, macrophage; Mono, monocyte. b, Left, UMAP visualization of T and ILC subsets across all locations. Right, distribution of lymphoid subsets (top) and cell numbers (bottom). NK, natural killer. c, UMAP visualization of Treg cell subsets across all locations (top left) and overlay on the UMAP plot of expression data for selected genes (middle; colours show relative expression). Right, distribution of Treg cell subsets (top) and cell numbers (bottom). d, Left, representative FACS plots of c-MAF versus TCF1 for Treg cells in tissue and MLN of Hh-colonized mice. Frequency of c-MAF+ (centre) and TCF1+ (right) Treg cells in tissue and MLN of SPF and Hh-colonized mice. e, Clonotype network analysis of TCRHh T cells and host clones by location (left) and cell phenotype (right). Each fully connected subnetwork represents a ‘clonotype cluster’ and each dot represents cells with identical receptor configurations. Clonotypes with fewer than two cells were filtered out for visualization. Prolif., proliferating. a–c,e, One sequencing run from six combined mice. d, Five SPF and six Hh-colonized individual mice, representative of two independent experiments. d, One-way ANOVA using a Tukey’s multiple comparisons test.
The T cell and ILC compartment comprises 16 transcriptionally distinct subsets (Fig. 2b). As expected, naive CD4+ and CD8+ T cells were mainly located in the MLNs and CP. Because photo-activation was successfully targeted to the T cell zone of each tissue, we did not detect T follicular helper cells in any compartment. As described in recent work, MHCIIhi type 3 ILCs (ILC3s) were identified in the MLN12,13,14, with a smaller population of MHCIImid/low ILC3s in the LA. Spatial compartmentalization of ILCs within the tissue showed that ILC3s were dominant in the LA and ILC2s were dominant in the LP (Fig. 2b and Supplementary Data 3b). The LP also contained a sizeable population of memory CD4 T cells that were distinct from the γδ T cells and T helper 17 (TH17) cells found in the MLN (Fig. 2b and Extended Data Fig. 5e). This heterogenous population contained cells that could be broadly classified as TH17, T helper 1 (TH1) and circulating memory populations (Extended Data Fig. 5f,g). Treg cells were enriched in both LP and LA at higher levels than other CD4 cell subsets (Fig. 2b and Extended Data Fig. 5h).
Subclustering of Treg cells revealed three subpopulations that were differentially distributed between secondary lymphoid organs and tissue (Fig. 2c). To highlight their distinct functional phenotype and spatial segregation, we refer to lymphoid-associated Treg cells as central Treg (cTreg) cells and tissue resident Treg cells as effector eTreg cells. cTreg cells express Tcf7 and Ccr732,33, tissue eTreg cells express the effector-associated molecules Ctla4, Maf, Tnfrsf4 and Il10, and proliferating Treg cells express histone genes (Fig. 2c and Extended Data Fig. 6a). Psuedotime analysis suggests a developmental relationship between these populations3,15 (Extended Data Fig. 6b,c). We sought to understand how the cTreg–eTreg dichotomy compared with previously described intestinal Treg cell subsets. Our data reveal Rorc, Gata3, Ikzf2 and Nrp1 expression across both cTreg cell and eTreg cell subsets (Fig. 2c). Although Nrp1 expression was higher in cTreg cells, it was not widely or exclusively expressed, suggesting that it would not qualify as a lineage defining marker. Expression of Gata3 and Rorc was almost always mutually exclusive in Treg cells (Extended Data Fig. 6d). Overlay of Maf expression suggests that both Gata3 and Rorc Treg cells are capable of upregulating the Maf-associated eTreg cell suppressor program (Extended Data Fig. 6d). Comparison of Rorc Treg cells in the MLN and LP again demonstrates differentially expressed Tcf7 (Extended Data Fig. 6e), supporting the idea that Rorc-expressing Treg cells in the MLN adopt a cTreg cell phenotype. Because unbiased clustering of the Treg cells separates cells on the basis of the cTreg–eTreg divide, this suggests that these developmental states represent overriding environmental imprinting based on interactions within the microniches that are dominant over the previously described ontological distinctions. The cTreg versus eTreg phenotype is also observed in specific pathogen free (SPF) mice on the protein level, suggesting that these distinct Treg cell phenotypes are not limited to the Hh setting (Fig. 2d).
To determine whether host and transferred cells were comparable for downstream analysis, we performed a T cell clonotype analysis. We detected TCRHh T cells (clonotype 206), which demonstrated that these cells become eTreg cells in the tissue and also represent the most expanded clone in any tissue (Fig. 2e and Extended Data Fig. 6g–h). TCRHh T cells could be found across all locations but were enriched in LA and LP (Fig. 2e and Extended Data Fig. 6h). Uniform manifold approximation and projection (UMAP) overlay of TCRHh T cells onto the total T cell (Extended Data Fig. 6g) or Treg cell (Extended Data Fig. 6f) pool confirmed TCRHh T cell distribution amongst the host clonotypes, not as a separate, unique cluster, supporting the idea that these cells can be viewed as representative of intestinal Treg cells. By focusing on clones that are present in the LP, we identified expanded host clones—for example, clone 1710 that also differentiates into eTreg cells within this microniche, and clone 1773 that represents proliferating Treg cells within the LP (Fig. 2e). We also found evidence of shared clonotypes within (for example, clones 17 and 703) and across tissues (for example, clone 854) in different differentiation stages. Clonotypes 34 and 406 support the prediction from the trajectory analysis (Extended Data Fig. 6b) that a clonotype present as a cTreg cell in a secondary lymphoid structure could further differentiate into an expanded eTreg cell population within the LA and LP. Analysis of differentially expressed genes within the eTreg cell populations across tissue microniches identified upregulation of Gzmb, Areg, Ccr2 and Itga4 in the LP and Cxcr4 in the LA34,35 (Extended Data Fig. 6i). This highlights the LP as a key site favouring optimal eTreg cell phenotype. Overlaying TCRHh T cells onto the heat map, we did not observe any phenotypic difference between the TCRHh T cells and their neighbouring cells within the microniches (Extended Data Fig. 6i). UMAP overlay onto the total Treg cell pool confirmed that TCRHh was located primarily within the LP eTreg cell cluster (Extended Data Fig. 6f). Owing to the small numbers of TCRHh T cells captured (Extended Data Fig. 6h), subsequent analysis combined host and TCRHh cells, as they are transcriptionally similar (Extended Data Fig. 6i). This further supports TCRHh as a representative, sentinel population within the T cell microniches.
After identifying spatially segregated cTreg and eTreg cells and enhanced eTreg suppressor function within the LP, we blocked IL-10, a key eTreg cell effector molecule, to determine the stability of the eTreg phenotype in the tissue. This approach is different to the standard Hh/anti-IL10R colitis model3 because in our model, the tissue responses to Hh are established before we perturb IL-10 signalling. We transferred TCRHh T cells into colonized hosts as before, allowing TCRHh T cells to migrate to the gut 7 days after transfer and adopt an IL-10+ Treg fate. We treated the hosts with FTY720 from day 8 to stop further migration from the secondary lymphoid organs and disrupted the IL-10 positive-feedback loop with an IL10Rα blocking antibody on day 10 after cell transfer (Extended Data Fig. 7a).
At 3 days after anti-IL10R treatment (13 days after TCRHh cell transfer), there was no significant difference between the two treatment groups in terms of the proportion or phenotypes of TCRHh Treg cells (Extended Data Fig. 7b). However, 7 days after anti-IL10R treatment (17 days after TCRHh cell transfer), there was an increase in local proliferation of TCRHh and host T cells, with approximately 60% of TCRHh Treg cells and TCRHh TH17 cells expressing Ki-67, which was not affected by FTY720, suggesting that these changes were generated locally (Extended Data Fig. 7c). Proliferating eTreg cells expressed similar amounts of FOXP3 and c-MAF compared with non-proliferating cells (Extended Data Fig. 7d,e), suggesting that this population is phenotypically stable and capable of local proliferation. Although we observed some increased cytokine expression, we did not observe overt inflammation in FTY720-treated or untreated conditions (Extended Data Fig. 7f–i). This does not rule out the possibility that critical tolerance-establishment steps occur in the MLN, but ongoing input from the MLN is not required to maintain tissue homeostasis. To determine which niche in the tissue supported Treg cell proliferation in this perturbed environment, we stained tissues with Ki-67, FOXP3 and Hoechst. This demonstrated Treg cell proliferation in the LP (Extended Data Fig. 7j,k), again supporting the LP as the key location for eTreg cell proliferation. These data suggest that although IL-10 is critical for initiation of tolerance, other eTreg cell suppressor functions can constrain the local response.
Myeloid compartmentalization
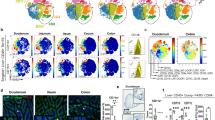
The compartmentalization of enhanced Treg cell function in the LP, as demonstrated by imaging and transcriptional studies, suggests that different APC populations may be stimulating and responding to eTreg cells in LP and LA36. Mapping of monocyte/macrophage and dendritic cell subsets across the four regions showed some clustering by region; however, it did not reveal a unique APC population (Fig. 3a). We did identify an enrichment of IL-1βhiCD103+SIRPα+ dendritic cells within LA, although IL-1β+ and CD206+ macrophages and IL-1β+ monocytes were most abundant in the LP (Fig. 3b). Immunofluorescence staining of SIRPα+ dendritic cells shows them densely populating the LA (Fig. 3c,e), whereas CD206 staining demonstrates the presence of CD206+ macrophages in the LP (Fig. 3d,e). Because a composite MHCII score suggests that all myeloid populations across compartments contain MHCIIhi cells, antigen-driven interactions may occur between any of these groups and eTreg cells (Fig. 3f).
a, Left, UMAP visualization of total myeloid subsets across all locations. Middle, UMAP of total myeloid populations by tissue microniche. Right, myeloid subset distribution (top) and total cell numbers at the indicated locations (bottom). b, log2-transformed fold difference of proportions of myeloid, T cells and ILCs in LA and LP. c, Representative CD11c and SIRPα immunofluorescence and DAPI staining in Hh-infected caecum tissue, showing an LA and surrounding LP. Scale bar, 50 μm. d, Representative CD206 immunofluorescence and DAPI staining in Hh-infected caecum tissue showing an LA and surrounding LP. Scale bars 50 μm. e, Densities of CD11c+SIRPα+ dendritic cells and CD206+ macrophages in the LA and LP microniches of Hh-infected caecum tissue. f, Violin plots of MHCII expression score across lymphoid and myeloid cells in the LP (top) and LA (bottom). Restricted to cell types with more than 30 cells per region. g, Relative mean expression of genes associated with STAT3 signalling and receptors in myeloid and lymphoid subsets in the LA and LP. Restricted to cell types with more than 30 cells per region. a,b,f,g, One sequencing run from four T cell compartments from six combined mice. c–e, Two independent experiments with n = 6 per group. Each dot represents data from one individual mouse. e, One-way ANOVA using Tukey’s multiple comparisons test.
Because IL-10 and other STAT3-activating cytokines are key to controlling homeostasis and inflammation in the intestine, we performed a search of STAT3-dependent cytokine genes across the most abundant myeloid and lymphoid subsets in the tissue. We found upregulated expression of the pro-inflammatory cytokine gene Il6 in IL-1β+CD103+SIRPα+ dendritic cells resident in the LA (Fig. 3g). By contrast, the LP was dominated by Il10, which is expressed primarily by eTreg cells, with some expression in CD206+ and IL-1β+ macrophage subsets (Fig. 3g). A small amount of Il27 was detected in the IL-1β+ macrophage population, specifically in the LP. Together these data suggest potential myeloid cytokine microniches that are capable of tuning STAT3 signals, potentially establishing inflammatory (LA) and anti-inflammatory (LP) niches within the intestinal tissue. We next examined expression levels of cytokine receptor genes. Moderate levels of Il6ra were detected across myeloid populations, and Il27ra expression was largely restricted to eTreg cells, CD4+ memory T cells and ILC2 populations. Il10ra was highly expressed on CD206+ and IL-1β+ macrophages and IL-1β+ monocytes, especially in the LP niche (Fig. 3g). Macrophage sensing of IL-10 is critical for gut homeostasis37,38, and our data support a potential positive-feedback loop in the LP between macrophages and eTreg cells. CD206+ and IL-1β+ macrophages respond to IL-10 and produce IL-10 and IL-2739,40, activating STAT3 and supporting the Maf program in eTreg cells in the LP.
Niche disruption in colitis
We aimed to extend our study to the entire intestinal tissue, validate our findings with a complementary method and include an inflammatory setting. Spatial transcriptomics is an unbiased approach to spatially map transcriptional signatures onto histological images of tissue sections41,42, enabling the inclusion of cells absent from the NICHE-seq pipeline. We processed frozen caecum and proximal colon tissue Swiss roll sections from mice treated with Hh only and anti-IL10R blockade at the time of infection to induce colitis for spatial transcriptomics using the Visium (10X Genomics) platform (Fig. 4a and Supplementary Data 5). The expression of cell cycle genes was increased across the tissue in the Hh/anti-IL10R setting, indicating a strong inflammatory response (Extended Data Fig. 8b). We integrated the annotations from our NICHE-seq data with data from Biton et al.30, Drokhlyansky et al.27 and Xu et al.15 and used the Cell2location pipeline43 to spatially map 102 transcriptionally distinct cell types onto the tissue sections (Fig. 4a). Manual annotation based on histology identified LAs and their overlying mRNA capture spots, distinct from the surrounding LP (Extended Data Fig. 8a). Non-negative matrix factorization (NMF) analysis identified 18 distinct transcriptional groups, covering all tissue microniches from microglia-associated (factor 5) to LA-associated (factors 3 and 15) (Extended Data Fig. 8c,d). Factor 2 includes colocalized eTreg cells, ILC2s and memory CD4 T cells that were previously identified in the LP by NICHE-seq (Extended Data Fig. 8c). Manual annotation of LAs enabled validation of NMF. Both techniques show similar trends of increased LA annotated tissue microniches in the context of colitis, probably reflecting development of LA into isolated lymphoid follicles (Fig. 4b). Example overlays of factor analysis, manual annotation and haematoxylin and eosin (H&E) staining demonstrate that the LA niche-associated cell signature (eMBCs, naive CD8s and lymphoid tissue inducer (LTi)-like ILC3s) overlap (Fig. 4c and Extended Data Fig. 8e). To understand whether the LP or LA niches are key to tolerance, we examined changes in IL-10 localization during inflammation. IL-10 was enriched in the LP (Extended Data Fig. 8f), as expected on the basis of NICHE-seq data (Fig. 3g). By comparing cell-type loadings across the LA and LP microniches in the Hh-only setting, we could identify IL-1βhiCD103+SIRPα+ dendritic cells, ILC3s and eMBCs in the LA, whereas more eTreg cells, CD206+ macrophages, ILC2s and IgA+ plasma cells were present in the LP, further supporting the cell enrichment in microniches identified through NICHE-seq analysis (Extended Data Fig. 8g). Expanding our analysis to the colitic Hh/anti-IL10R setting, we observed enrichment of some cell types, including ILC2s, in the LP; however, cells previously restricted to the LA such as IL-1βhiCD103+SIRPα+ dendritic cells and ILC3s could be found in the LP (Extended Data Fig. 8g). Comparing the log-transformed odds ratio (OR) of each cell type between LA and LP, we could identify cells that were significantly enriched within the LA (Fig. 4d, purple bars representing adjusted P value < 0.05). To include more spots in the analysis, a comparison of LA versus the rest of the tissue showed similar results (Extended Data Fig. 8h,i). As expected, this more statistically rigorous analysis showed that IL-1βhiCD103+SIRPα+ dendritic cells, ILC3s and eMBCs were significantly enriched in the LA of the Hh-only setting (Fig. 4d). The overall magnitude of enrichment quantified by log(OR) across all populations was lower during colitis, and IL-1βhiCD103+SIRPα+ and RORγt+MHChi ILC3s were no longer significantly enriched. This is consistent with cells trafficking from LA to LP, as previously observed with ILC3s44, and potential recruitment of additional cells into this microniche, disrupting the tolerogenic niche. Analysis of immunofluorescent staining of SIRPα+ dendritic cells in the Hh/anti-IL10R condition, showed similar distribution between the LA and LP in the context of colitis (Fig. 4e,g), which mirrors the findings from the cell-loading analysis. Similarly, staining of CD206+ macrophages showed no significant difference between LA and LP in colitis (Fig. 4f,g). We used analysis of the cell density from immunofluorescent staining to validate the ratio analysis in Fig. 4d. This demonstrated that the SIRPα+ dendritic cells, which are significantly enriched in the LA at steady state, are no longer significantly enriched in colitis (Fig. 4h). Co-occurrence analysis in Hh and Hh/anti-IL10R settings showed several immune cell populations that correlate with each other within Visium spots. By comparing the change between homeostasis and colitis, we can identify co-occurrences that significantly change in the context of inflammation. The most significantly increased co-occurrence was between CD103+SIRPα+ dendritic cells and eTreg cells, again suggesting that these dendritic cells may disrupt the homeostatic interactions within tissue (Extended Data Fig. 9a).
a, Schematic of the setup and analysis of the spatial transcriptomic experiment. b, Top, relative abundance of LA in Hh-infected mice versus Hh/anti-IL10R mice by NMF analysis (top) and manual annotation (bottom). c, H&E staining of Hh-infected tissue. Localization of LA-associated factor 15 by NMF decomposition as defined in Extended Data Fig. 8c (top left) and manual annotation (top centre and right) in the Visium RNA capture spots of a representative sample. Localization and normalized cell-type abundance of eMBCs (bottom left), naive CD8+ T cells (bottom middle) and LTi-like ILC3s (bottom right) in a representative LA. Scale bars, 200 μm. d, Cell-state enrichment ordered by cell lineage in the manually annotated LA versus LP spots in Hh-infected (top) and Hh/anti-IL10R (bottom) mice. Statistically significant enrichments (chi-square test, adjusted P value (Padj) < 0.05) are shown in magenta. Data are log2OR value ± s.d. progen. progenitor; TA, transit amplifying; GC, germinal centre; GC.BC/DZ-pre-mem, germinal centre B cell/dark zone and pre-memory; PC, plasma cell. e, CD11c and SIRPα immunofluorescence and DAPI staining in Hh/anti-IL10R mice, showing LA and surrounding LP. Scale bar, 100 μm. f, CD206 immunofluorescence and DAPI staining in Hh/anti-IL10R mice, showing LA and surrounding LP. Scale bar, 100 μm. g, Density of CD11c+SIRPα+ dendritic cells and CD206+ macrophages in LA and LP of Hh/anti-IL10R mice. h, Ratio of SIRPα+ dendritic cell and CD206 macrophage cell densities in LA vs LP in Hh and Hh/anti-IL10R mice. a–d,i, Four individual mice per group sequenced from two Visium slides. e–h, Two independent experiments with n = 6 per group. Each dot represents one mouse. g,h, One-way ANOVA using Tukey’s multiple comparisons test with single pooled variance.
eTreg cell–macrophage interactions in LP
eTreg cells and IL-10 are more enriched in LP at homeostasis (Extended Data Fig. 8f,g), in contrast to colitis, suggesting that there are likely to be unique interactions that are lost in the context of inflammation, where dendritic cells dominate the LP. To understand which signals are important for bringing together cells of interest and contributing to local cytokine delivery, we performed CellPhoneDB analysis45 using NICHE-seq data from the Hh setting for each tissue microniche. Chord diagrams of significant interactions between any cell pair in the LP show that eTreg cells are capable of interacting with all cell populations within the niche, with many potential interactions across APC subsets (Extended Data Fig. 9b). This fits with their dynamic nature, as observed previously with intravital microscopy (Supplementary Video 3). The interaction diagram for LA shows that eTreg cells have more interactions with IL-1β+CD103+SIRPα+ dendritic cells in particular (Extended Data Fig. 9b).
CellPhoneDB analyses of unique cell-pair interactions with a more stringent P < 0.01 in the LA and LP catalogue key predicted interactions between cells in each tissue microniche (Fig. 5a). In addition to differences in interactions with APCs, the eTreg cells in the LP had the strongest ICAM1–ITGAL interactions with CD4 memory T cells and LTBR–LTB interactions with ILC2s, whereas eTreg cells in the LA could interact with ILC3s through ICAM1–ITGAL (Fig. 5a). These eTreg cell–ILC2 interactions observed across experimental analyses (Fig. 2b and Extended Data Fig. 8c,g) provide a potential source of IL-2 for the eTreg cells within the LP niche (Extended Data Fig. 9c). Of particular interest are the chemokine–chemokine receptor and integrin pairs that bring APCs and eTreg cells together. Within the LP, eTreg cells have the strongest CCR2–CCL8, PTPRC–MRC1, CSF1–CSFR1 and VCAM1–A4B7 pathway interactions with CD206+ macrophages, suggesting that these cells may have a dominant role beyond TCR engagement (Fig. 5a). Within the LA, the CCR2–CCL8, PTPRC–MRC1 and CD72–SEMA4D interactions connect eTreg cells with IL-1β+ macrophages, and CXCR3–CXCL9 and ICAM1–ITGAL interactions connect eTreg cells with LA dendritic cell populations (Fig. 5a). On the basis of the unbiased CellPhone analysis we further analysed CCR2 expression in the T cell compartment, which is highest in eTreg cells and some TH17 cells (Extended Data Fig. 9d,e). The ligands, CCL7 and CCL8, are most highly expressed by CD206+ macrophages, further supporting a potential key role for eTreg cell–CD206+ macrophage interactions in the LP niche (Extended Data Fig. 9e). Compartmentalization of CCR2 in LP and CXCR3–CXCR4 in LA is further validated by expression levels and receptor–ligand analyses in spatial transcriptomics data (Extended Data Fig. 9f,g).
a, CellPhoneDB analysis of receptor–ligand interactions in LP (top left) and LA (top right), restricted to cell types with more than 30 cells per region and limited to unique pairs with P < 0.01. MHCII interactions are excluded. Schematic summarizing the most relevant cell–cell interactions (bottom). b, Two-photon in vivo live imaging of TCRHhCD2DsRedIl10GFP T cells transferred into Hh-colonized hosts labelled in vivo with CD206 and F4/80 fluorescent antibodies. c, Representative image of donor TCRHhCD2DsRedIl10GFP T cells in the LP. Arrowheads indicate Il10− TCRHh (red) and Il10+ TCRHh (yellow) T cells. Scale bar, 20 μm. d, Average track speed of Il10+ and Il10− TCRHh T cells in the LP. e, Track displacement length of Il10+ and Il10− TCRHh T cells in the LP. f, Sequential video stills showing an Il10+ TCRHh T cell moving from one CD206+ cell to another in the LP. Scale bars, 10 μm. g, Distance from Il10+ and Il10− TCRHh to the nearest CD206+ cell in the LP. a,b, One sequencing run from six combined mice. d–h, Representative images and combined data from six individual mice over two independent imaging experiments. d,e,g, Mann–Whitney test.
On the basis of the CCR2–CCL8 axis highlighted in the CellPhoneDB data and differential expression of CCR2 in LP eTreg cells (Fig. 5a and Extended Data Figs. 6i and 9d–g), we hypothesized that eTreg cells interact directly with CD206+ macrophages in the LP. To test this, we performed in vivo two-photon live imaging of TCRHhCD2DsRedIl10GFP T cells transferred into Hh-colonized hosts and labelled in vivo with F4/80 and CD206 antibodies (Fig. 5b). Imaging the LP showed Il10+ TCRHh T cells intimately associated with CD206+F4/80+ macrophages, in contrast to their Il10− TCRHh T cell counterparts (Fig. 5c). Despite similar cell track speeds, Il10-GFP+ TCRHh cells exhibited greater displacement (Fig. 5d,e). This suggests that Il10-GFP+ TCRHh T cells are highly motile over large areas of LP, whereas Il10-GFP− TCRHh T cells are motile within a more confined area. Analyses of time-lapse images show Il10-GFP+ TCRHh cells moving from one CD206+ macrophage to another (Fig. 5f and Supplementary Video 4). The result of motile eTreg cells interacting with CD206+ macrophages in the LP is that the distance from any Il10-GFP+ TCRHh T cell to a CD206+ macrophage is significantly smaller compared with Il10− TCRHh cells, with about one-third of cells being in contact with a CD206+ macrophage at one time (Fig. 5g).
Discussion
In this study, we follow the natural history of TCRHh as a model of microorganism-reactive Treg cells as they acquire and maintain immune regulatory function in the intestine. By examining cells in anatomical microniches, we have revealed the importance of key interactions to enhance eTreg cell antigen stimulation and effector function. The cTreg–eTreg differentiation trajectory identified using TCRHh T cells and mirrored in endogenous Treg cells suggests that overriding environmental imprinting based on interactions within microenvironments drives Treg phenotype, overlaying this paradigm on the Gata3–Rorc dichotomy. Even among eTreg cells, the LP—and not the LA—is the site of enhanced eTreg cell function, including the production of AREG, GZMB and IL-10.
The LP is unique in its cellular makeup, supporting tolerance at steady state. This microniche is inhabited by eTreg cells and a diverse set of APCs, including CD103+ dendritic cells, CD206+ macrophages, IL-1β+ macrophages, and monocytes that express the highest levels of IL10R, making them prime targets for IL-10 produced by eTreg cells. Once this tolerogenic niche is established, eTreg cells can proliferate within this microniche to control pathology. However, the LP niche is perturbed in the context of inflammation, with several cell types including CD103+SIRPα+ dendritic cells previously confined to the LA, recruited into the LP, again pointing to this tissue as key to mucosal tolerance. Within the LP, IL-10-producing Treg cells are motile and participate in serial interactions with CD206+ macrophages. eTreg cells and CD206+ macrophages express several molecules capable of potentiating these interactions, including attraction through CCR2–CCL8, adhesion through VCAM1–α4β7 and CD45–CD206 and immune activation and control through TCR–MHC and IL-10–IL10R.
This work highlights the importance of the eTreg cell program, and the motility and stability of eTreg cells, which are uniquely poised to interact with cells across a large barrier surface. By characterizing their niche in addition to their cellular phenotype, we have uncovered several pathways that may be enhanced or disrupted to control pathology. Information gained from the study of microniches can be used for targeted interventions, cell therapy and vaccination strategies to support eTreg cell recruitment, activation, differentiation, survival and function in the inflammatory niche of the intestine and other organs.
Methods
Mice
C57BL/6 J (B6) and Foxp3GFP, Rag1−/−, CD2DsRed, Foxp3CD2Il10GFP, UbPA-GFP and Nur77GFP mice were bred and maintained under specific pathogen free conditions in an accredited animal facility at the University of Oxford. Lta−/− mice were purchased from Jackson Laboratories. HH7-2tg TCR transgenic mice, referred to as TCRHh here, were provided by D. R. Littman. TCRHh mice were bred with CD2DsRed mice and either Foxp3CD2Il10GFP or Nur77GFP mice to generate TCRHhCD2DsRedIl10GFP or TCRHhCD2DsRedNur77GFP, respectively. TCRHh mice were bred to Rag1−/− to generate TCRHhRag1−/− mice. UBPA-GFP mice were bred to CD2DsRed and TCRHh mice to generate CD2DsRedUbPA-GFP and TCRHhCD2DsRedUbPA-GFP, respectively.
Mice were free of Helicobacter spp. and other known intestinal pathogens, were age- and sex-matched and between 6 and 12 weeks old. Animals were randomly assigned to experimental group, and cages contained mice of all different experimental groups. All experiments were conducted in accordance with the UK Scientific Procedures Act of 1986, and by persons holding a personal license. The project licence governing the mouse studies was reviewed by the University of Oxford’s Animal Welfare and Ethical Review Board and approved by the Home Office of His Majesty’s Government.
No statistical methods were used to predetermine sample size. Sample sizes were based on previous similarly designed experiments from our research group. The spatial transcriptomics experiment included four mice per group to balance statistical power with cost. For other experiments we aimed for a minimum of five mice per experimental group. Exact mouse numbers for each experiment are included in the figure legends. Mice were assigned to different experimental groups at random. Mice were co-housed and littermate when possible. Each cage contained all treatment conditions. Animal studies were not blinded. Histopathology scoring was conducted by two independent assessors, one of whom was blinded.
Flow cytometry
Mouse cells were stained with combinations of the following monoclonal antibodies, all purchased from Biolegend, Invitrogen, or eBioscience: CD4 (RM4-5), TCRβ (H57-957), CD45.1 (A20), CD45.2 (104), CD11c (N418), CD11b (M1/70), anti-human CD2 (TS1/8), CXCR5 (L138D7), PD-1 (J43), FOXP3 (FJK-16s), RORγt (Q31-378), Ki-67 (SolA15). Dead cells were excluded using efluor 780 fixable viability dye (eBioscience). For transcription factor staining, cells were stained with surface markers prior to fixation and permeabilization using the FOXP3 staining buffer kit (eBioscience) according to manufacturer instructions.
Immunofluorescence staining
Swiss-rolled caecum tissues were fixed overnight at 4 °C in PLP buffer (1% paraformaldehyde, l-lysine 0.2 M pH 7.4 and 32 mg NaIO4). Then, tissues were dehydrated in 20% sucrose for at least 4 h at 4 °C and embedded in OCT compound (Avantor). Seven-micrometre cryosections were rehydrated, blocked and permeabilized with PBS, 1% goat serum, 1% BSA, 0.3 M glycine, 0.3% Triton X-100 for 1 h at room temperature. Sections were stained with the following antibodies: Alexa Fluor 488 anti-mouse CD172a (SIRPα) (clone P84, 5 μg ml−1 Biolegend), Alexa Fluor647 anti-mouse CD11c (clone N418, 5 μg ml−1 Biolegend) or FITC/Alexa Fluor 594 anti-mouse CD206 (clone C068C2, 5 μg ml−1 Biolegend) and anti-mouse CD64 (clone X54-5/7.1, 4 μg ml−1 Biolegend). Sections were stained overnight at 4 °C. Before imaging, nuclei were counterstained with Hoechst. Images were acquired using Zen Blue software on a ZEISS 980 Airyscan inverted microscope equipped with a motorized stage. Diode laser lines were used for excitation: violet (405 nm), blue (488 nm), yellow (514 nm) and red (639 nm). All images were acquired with a 25× (NA 0.8) LD LCI Plan-Apochromat oil-immersion objective.
Colon tissue was embedded in OCT (Tissue-Tek) as Swiss rolls and sectioned at 7 μm. Slides were fixed with 3.7% formalin (Merck) and blocked with 10% donkey serum (Sigma Aldrich) and 1% Fc block (eBioscience) in permeabilization buffer (Foxp3/Transcription factor staining buffer set, eBioscience). B220 (RA3-6B2), CD4 (RM4-5), MHC Class II (M5/114.15.2), gp38 (8.1.1), IgD (11-26 c.2a) and BCL6 (IG191E/A8) (all Biolegend) were stained overnight in blocking serum.
Isolation of lymphocytes from spleen, lymph node and intestinal tissue
Intestinal tissues were washed twice in RPMI (Sigma Aldrich)/10%FCS/5 mM EDTA at 37 °C with agitation for 25 min to remove epithelial cells. CP and OLS were removed under 40× bright-field microscopy using a scalpel and a 16G needle and syringe, respectively. Remaining colon and caecum tissue, OLS and CP were digested for 40 min at 37 °C with agitation in RPMI/10% FCS/15 mM Hepes with 100 U ml−1 collagenase VIII (Sigma Aldrich) and 20 mg ml−1 DNase I (Sigma Aldrich). Leukocytes from colon and caecum tissue were recovered at the interface of a 40/70% Percoll gradient (Fisher Scientific).
Spleens and MLNs were mechanically disrupted, and splenic red blood cells were lysed with ACK lysis buffer.
Peripheral blood was collected by cardiac puncture and red cells were lysed with ACK lysis buffer.
Hh culture and oral gavage
Hh NCI-Frederick isolate 1 A (strain 51449) was grown on blood agar plates containing 7.5% laked horse blood (Thermo Scientific) and Skirrow Campylobacter supplement (Oxoid) under microaerophilic conditions at 37 °C with agitation. Cultures were expanded for 48 h in Tryptone Soy Broth (TSB, Fisher) containing 10% FCS (Gibco) and the above antibiotics. The concentration of bacteria was determined by optical density (OD) analysis at 600 nm. Mice were fed 1 × 108 colony-forming units of Hh (equivalent to 1 OD unit) by oral gavage using a curved 22G needle for a total of 2 doses 24 h apart.
Lymph node lymphocyte egress blocking experiment
Host mice were treated every 24 h with 1 mg kg−1 of the sphingosine-1-phosphate antagonist, Fingolimod (FTY720, Sigma Aldrich) via intraperitoneal injection at the indicated timepoints after naive TCRHh cell transfer.
Sorting and adoptive transfer of naive TCRHh T cells
Naive T cells were isolated from TCRHh mice splenocytes and sorted by flow cytometry as CD45+CD3+CD4+CD44lowCD62LhiVβ6+ (Extended Data Fig. 1d), with up to 2% contamination with nTreg cells. All monoclonal antibodies were purchased from Biolegend or eBioscience: CD3 (145-2C11), CD11b (M1/70), CD11c (N418), B220 (RA3-6B2), CD62L (MEL-14), TCRVβ6 (RR4-7), CD44 (IM7), CD45 (30-F11), CD4 (RM4-5). Sorted cells (2 × 103 or 5 × 104) were injected by intravenous injection into the tail vein for flow cytometric or in vivo live imaging respectively.
In vitro co-culture
Bone marrow stem cells were extracted from wild-type mouse femurs and cultured for 7 days in RPMI (Sigma) supplemented with 1% penicillin-streptomycin (Sigma), 10% FCS (Life Technologies), 1% Glutamax (Invitrogen) and 20 ng ml−1 mouse GM-CSF (Peprotech). Bone marrow-derived dendritic cells (BMDCs) were plated at a density of 1 × 104 cells per well overnight. Hh peptide (1 mg ml−1, Genscript) was added 1 h prior to plating 1 × 105 sorted naive TCRHhNur77GFP T cells in RPMI/10% FCS/1% Glutamax/1% penicillin-streptomycin and 50 mM β-mercaptoethanol (Life Technologies). Anti-mouse I-A/I-E antibody was added at 4, 12, 24, 48, 72 and 96 h after the plating of TCRHhNur77GFP T cells.
Two-photon microscopy
Mice were anaesthetized with isoflurane, the caecum exposed and immobilized with a suctioning imaging window46. Samples were illuminated with 910 nm <70 fs pulsed light from Mai-Tai laser and collected using a 20× water-dipping lens and the spectral detector of a Zeiss 880 multiphoton microscope (Carl Zeiss). Images were linearly unmixed using the Zen software (Carl Zeiss) to separate autofluorescence, collagen, eGFP, DsRed and Texas-red dextran based on single-colour controls. All in vivo live images of the LA and LP were performed in the caecum due to ease of access.
Intravital videos were drift-corrected based on mucus or collagen signal. Images were smoothed using a Gaussian filter for display.
Image analysis
Intravital microscopy images were analysed using Imaris 9 (Bitplane). Following unmixing, autofluorescence was subtracted from DsRed and GFP channels. GFP+ and DsRed+ cells were marked using the Surface Creation Wizard, and their co-expression and location within LP and LA compartments were recorded.
Immunofluorescence images were analysed using Imaris 10.0 (Bitplane). LAs were defined based on DAPI staining showing nuclear density and surrounding epithelial morphology, allowing for unbiased region selection. LP was chosen based on location and cellular density based on DAPI stain so that each region of interest collected and analysed for LA contained at least six LP surfaces of matching size. Surfaces for each cell type of interest were created using CD11c+ for BMDCs, CD206+ for CD206+ macrophages using the Surface Creation Wizard, which was applied to all images collected with the same conditions. CD11c+ surfaces were further subdivided based on median SIRPα staining levels within the surface.
Photo-activation
CD2DsRedUbPA-GFP hosts were colonized with Hh on day 0. Seven days later 50,000 naive TCRHhCD2DsRedUbPA-GFP cells were transferred into the Hh-colonized CD2DsRedUbPA-GFP hosts so both host and donor cells were photo-activatable. On day 21 the MLN and caecum were removed from 6 mice. The caecum was shaken at 37 C in RPMI + BSA + EDTA for 40 min and 20 min to remove the epithelium. The CP was separated from the rest of the tissue, and the remaining tissue was divided into one third without photo-activation as a negative control for FACS gating, one third for LA photo-activation, and one third for LP photo-activation.
Samples were maintained in RPMI + BSA+Hepes on ice in the dark for the duration of the experiment with cold media flowed over the tissue during photo-activation. A Zeiss 880 upright multiphoton microscope (Carl Zeiss, Germany) fitted with two tunable lasers (Mai-Tai tuneable BB laser 710–990 nm, pulse width <80 fs and Mai-Tai tuneable 690–1040 nm, pulse width <70 fs) and a 20× water-dipping objective was used for tissue photo-activation. The microscope was set to dynamically unmix GFP, DsRed, and collagen based on pre-collected single-colour controls. Samples were imaged with 910 nm light to identify regions of interest based on CD2DsRed. MLN and CP T cell zones were defined as the densest T cell regions without gaps to exclude B cell zones. LA regions were defined as a cluster of cells with a diameter of at least ten CD2DsRed cells. LP region was defined as a region containing CD2DsRed cells distal to LA. After ROI definition, the second laser was turned on at 740 nm while imaging live. GFP photo-activation was observed dynamically to ensure sufficient photo-activation without toxicity. For each mouse 3–10 regions were photo-activated for each tissue microniche. Each photo-activation region comprised approximately 40,000 µm3 of tissue (70 µm diameter × 10 µm depth).
After photo-activation, tissues were minced and digested for 30 min in RPMI+Hepes with 100 U ml−1 collagenase VIII (Sigma Aldrich) and 20 mg ml−1 DNase I (Sigma Aldrich). The digested tissue from the six mice was pooled per tissue microniche into one sample. Isolated cells were stained with efluor 780 fixable viability dye (eBioscience). Photo-activated GFP+ cells were sorted using a four laser BD FACSAria III based on gates defined by unactivated samples from the same mice (Extended Data Fig. 5b).
Single-cell RNA library construction and sequencing
For scRNA-seq experiments, the Chromium Single Cell 5′ version 2 reagent kit and Chromium Single Cell Mouse TCR Amplification Kit (10x Genomics) were used. Sorted cells were loaded onto each channel of the Chromium Chip K following the manufacturer’s instructions and the chip was inserted in the Chromium Controller for droplet encapsulation. cDNA synthesis, amplification, gene expression (GEX) and targeted TCR was performed on single cells, according to the manufacturer’s protocol (CG000331). Sequencing was performed on the Illumina Novaseq 6000 system. Gene expression libraries were sequenced at a targeted depth of 50,000 reads per cell, using the following parameters: Read1: 28 bp i7:10 bp, i5: 10 bp, Read2: 98 bp. TCR libraries were pooled at a ratio of 1:10 with the GEX libraries and sequenced at a target depth of 5,000 reads per cell.
scRNA-seq analysis
Pre-processing of 10x Genomics scRNA-seq and scTCR-seq data
scRNA-seq raw sequencing data were processed using the CellRanger “multi” software (version 6.1.1, 10x Genomics) with the mm10 2020-A mouse reference genome (official 10X mouse pre-built reference). Single-cell TCR-sequencing (scTCR-seq) data were aligned and quantified using the CellRanger ‘multi’ software (v.6.6.1) and the reference vdj_GRCm38_alts_ensembl-5.0.0 was used with default settings.
Quality control and processing of scRNA-seq data
Data pre-processing was performed using the ScanPy workflow (v. 1.8.1)47. ScanPy (v. 1.7.1), Anndata (v. 0.7.5), Pandas (v.1.2.3), NumPy (v.1.20.1), and Python (v.3) were used to pool single-cell counts and conduct downstream analysis. For each run, SoupX algorithm48 was run with default parameters to remove ambient mRNA from the count matrix. Doublet detection was performed using the Scrublet algorithm (https://github.com/AllonKleinLab/scrublet49) with percolation step, as previously described50. Additional doublet exclusion was performed throughout downstream processing based on unexpected co-expression of canonical markers, such as Cd3d (T cells) and Cd19 (B cells). Cells with fewer than 1,000 unique molecular identifier (UMI) counts, fewer than 600 detected genes and more than 15% mitochondrial reads were excluded from downstream analysis. Genes were filtered out for expression in less than three cells. Gene expression for each cell was normalized (scanpy.pp.normalize_total, scaling factor 10,000) and log-transformed (scanpy.pp.log1p). Downstream analyses included variable gene detection (scanpy.pp.highly_variable_genes) and data feature scaling (sc.pp.scale). Cell cycle score was calculated using the expression of the cell cycle genes in Supplementary Table 1. Cell cycle score, UMI counts, the percentage of mitochondrial reads, and the percentage of Ig reads (calculated based on the genes provided in Supplementary Table 1) were regressed out during scaling the data. Dimensionality reduction (scanpy.tl.pca, based on highly variable genes) and Leiden-graph-based clustering (scanpy.tl.leiden, with clustering resolution manually adjusted, 0.3–1.5) were carried out. Cell lineages were annotated on the basimarker gene expression for each cluster (sc.tl.rank_genes_groups, method = ‘wilcoxon’).
Cell-type annotation with CellTypist
CellTypist is a cell-type database, server and pipeline for automatic annotation of scRNA-seq developed at Teichman lab (https://github.com/Teichlab/celltypist, https://www.celltypist.org). To assemble a mouse intestinal reference dataset, scRNA-seq data were collected from eight publications covering different cell lineages from small and large intestine as well as one additional dataset of sorted B cells from spleen, to cover in detail the annotation of germinal centre B cell populations (Supplementary Data 2).
For each dataset, the raw count matrix was downloaded along with the accompanying cell meta information. After removing trivial cell types annotated by the original studies (for example, ‘doublets’ and ‘unresolved’), a total of 171,271 cells were obtained representing all major cell populations in the mouse gut. Cell-type names from different datasets were next standardized to achieve a common scheme of nomenclature. Specifically, the similarity of transcriptomes between each pair of cell types across datasets was examined and the two cell types were merged only if they corresponded to a single cell type (for example, ‘enteroendocrine cell’ from the Tabula Muris was renamed to ‘enteroendocrine’ as was used in Biton et al.30). Finally, after in-depth inspection, 126 cell types were harmonized from the eight datasets. A CellTypist model then was created based on logistic regression classifiers, as described in detailed48. The model is publicly available at https://celltypist.cog.sanger.ac.uk/models/Mouse_Gut_Casado/v2/Adult_Mouse_Gut.pkl. Cell identities were predicted using the resulting model, followed by manual curation. Cells from each lineage were further subclustered and Leiden clustering was repeated for fine-grained annotation of the cell types and states. A description of cell-type annotations for each lineage is provided in Extended Data Fig. 5c and Supplementary Data 3 and 4. The differentially expressed genes for the cell types in each lineage can be found in the Supplementary Table 1.
For prediction on cycling regulatory T cells (prolif. Treg cells), eTreg and cTreg cells were used as a training reference. The model was built applying default parameters, and prediction was performed without majority voting.
scTCR-seq downstream analysis
The Python package scirpy (v. 0.12.2)51 was used to extract the V(D)J sequence information from the CellRanger output file filtered_contig_annotations.csv. Productive TCRαβ chains were determined using the scirpy.tl.chain_qc function, and cells without V(D)J data or with two pairs of productive TCRαβ chains were removed from the analysis. Clonotypes were defined with the function scirpy.tl.define_clonotypes based on the CDR3 nucleotide sequence identity and the V-gene usage for any of the TCR chains (either VJ or V(D)J need to match). For cells with dual TCRα or TCRβ chains, any of the primary or secondary receptor chain matching was considered for the clonotype definition. Clonotype networks were constructed using the pp.ir_dist function to compute distances between CDR3 nucleotide sequences (using identity as a metric) and tl.define_clonotype_clusters function to designate the clonotype clusters, removing the clonotypes with less than two cells. The TCR metadata were combined with the transcriptome data for downstream analysis and comparison of different T cell populations. Hh-specific TCR data were retrieved from Xu et al.15, and TCR sequences were obtained from https://www.ncbi.nlm.nih.gov/nuccore and mapped using the IMGT/V-QUEST alignment tool52. Hh.7-2 transgenic TCR (tgTCR) clonotypes were identified by expression of TRAV9-1/TRBV19 gene segments and the TCRβ CDR3 amino acid sequence, including those clonotypes with missing TCRα chain.
RNA velocity analysis
RNA velocity analysis53 was performed using the scVelo (v.0.2.4) package [10]. RNA velocity was estimated by distinguishing unspliced and spliced mRNAs using the velocyto package (v.0.17) (https://velocyto.org/velocyto.py/54). Data were subclustered on Treg cell, filtering out the subsets with fewer than ten cells per gut region (that is, excluding eTregs_MLN, eTregs_CP, cTregs_LA, cTregs_LP and Prolif-Tregs_CP). The dataset was then merged with the velocyto output (merged loom files) and pre-processed for detection of minimum number of counts, filtering and normalization (scvelo.pp.filter_and_normalize). The functions scvelo.pp.moments, scvelo.tl.velocity and scvelo.tl.velocity_graph were used to compute velocities using the stochastic mode in scVelo. The function scvelo.pl.velocity_embedding_stream was used to project the velocity information onto the UMAP. To test which genes have cluster-specific differential velocity expression and visualize selected genes, the functions scvelo.tl.rank_velocity_genes and scvelo.pl.velocity were applied. Velocity pseudotime was calculate with the function scvelo.tl.velocity_pseudotime.
Cell-type scoring
A list of mouse genes involved in the MHCII complex (Supplementary Table 2) was used for surface MHCII scoring. Cells were scored using the scanpy.tl.score_genes function according to the expression values of all genes.
Cell–cell communication analysis
The CellPhoneDB55,56 Python package (v.3 .0) was used to infer putative cell–cell interactions. The scRNA-seq dataset was split by gut region and cell types with <30 cells in a given region were filtered out. Human-mouse orthologue genes were retrieved using the biomaRt package57, and only one-to-one orthologous genes were considered. CellPhoneDB was applied on the normalized raw counts and fine cell-type annotations of myeloid, T cells and ILCs from LA and LP (separately for each gut region), using default parameters. To identify the most relevant interactions, specific interactions of Treg cells with myeloid cells and T cells/ILCs were selected and filtered for the ligand–receptor pairs that were significant (P ≤ 0.01) and ‘curated’. The selected interactions were plotted as expression of both ligands and receptors in relevant cell types. The ktplots R package (https://github.com/zktuong/ktplots/tree/plot_cpdb3; https://doi.org/10.5281/zenodo.5717923) was used to visualize the significant interactions per cell-type pair using a chord diagram.
Fresh frozen Visium sample preparation
Caecum and proximal colon tissue from Hh-infected and Hh/anti-IL10R treated mice were removed, cut longitudinally and cleaned of stool with cold phosphate buffered saline (PBS). The tissue was positioned luminal side up, and rolled into a Swiss roll from the caecum to the proximal colon. The tissue was placed into a histology plastic cassette and snap frozen for 1 min in dry-ice-cooled isopentane. The frozen tissue was embedded in OCT on dry ice and stored at −80 °C. The samples were selected based on tissue morphology and orientation (H&E-stained sections) and RNA integrity number, obtained using High sensitivity RNA ScreenTape system, (Agilent 2200 TapeStation). OCT blocks were sectioned at 10 μm thickness in a −20 °C cryostat (Leica CX3050S) at 10 μm, and transferred onto a 6 mm2 capture area on a Visium 10X Genomics slide. Visium spatial tissue optimization was performed, and an optimal permeabilization time of 24 min was selected. The Visium slides were processed according to manufacturer’s instructions, before fixing and staining H&E for imaging. H&E-stained slides were imaged at 40× on Hamamatsu NanoZoomer S60. After transcript capture, sequencing libraries were prepared according to the 10X Genomics Visium Spatial Transcriptomic protocol and sequenced on the Illumina Novaseq 600 system.
Visium spatial transcriptomics data analysis
10x Genomics Visium sequencing data processing
After sequencing, 10x Genomics Visium spatial samples were aligned to the mouse transcriptome mm10 2020-A reference (as the scRNA-seq samples) using 10x Genomics SpaceRanger version 2.0.0. and exonic reads were used to produce mRNA count matrices for each sample. SpaceRanger was also used to align paired histology images with mRNA capture spot positions on the Visium slides. A custom image-processing pipeline was used for alignment of Visium slides and identification of the spots contained in the tissue, as described in ref. 58. Spots with fewer than 500 UMI counts, and more than 15% mitochondrial genes were removed from the analysis. Data from different samples were concatenated and SCVI was used for batch correction59.
Spatial mapping of cell types using cell2location
To spatially map intestinal cell types defined by single-cell transcriptomics in the Visium data we used cell2location41. First, to obtain a complete single-cell reference of cell types and cell states in the mouse intestine we integrated our NICHE-seq data with 3 publicly available datasets of intestinal epithelial cells30, immune cells15 and enteric nervous system27 (Fig. 5a). Redundant cell annotations across different datasets were harmonized and curated manually. This scRNA-seq reference (untransformed and unnormalized mRNA counts) was then used in the cell2location pipeline, as described in detail previously43. In brief, reference signatures of cell states (63 cell populations) were estimated using a negative binomial regression model provided in the cell2ocation package. The inferred reference cell-state signatures were used for cell2location mapping that estimates the abundance of each cell state in each Visium spot by decomposing spot mRNA counts. The cell2location spatial mapping was done separately for Hh and Hh/anti-IL10R sections. The paired H&E images were used to determine the average number of cells per spot in the tissue (set to 30) and used as a hyperparameter in the cell2location pipeline. Cell-state proportions in each Visium spot were calculated based on the estimated cell-state abundances.
Two methods were used to identify cellular microenvironments in the tissue: manual annotation and conventional NMF analysis. Regions for manual annotation were defined based on H and E images. LA were defined by cellular density, whereas LP regions included histologically distinct areas both proximal and distal to the LA. NMF, implemented in the cell2location pipeline, was performed on cell abundance results by cell2location on each condition separately (Hh and Hh/anti-IL10R). The NMF model was trained for a range of factors and tissue zones (number of factors: n_fact) N = {5,…,30} and the decomposition into 18 factors was selected as a balance between segmenting relevant tissue zones (muscle compartment, lymphoid structures, lymphatics) and over-separating known zones into several distinct factors (Extended Data Fig. 8c).
Cell-state spatial enrichment analysis
Spots containing lymphoid aggregates and adjacent LP were manually annotated using the paired histology images of the spatial data in the 10x Genomics Loupe software. Cell-state proportions per spot were calculated based on the estimated abundances from cell2location and cell-state enrichments (odds ratio) in each manually annotated region were calculated as described60. In brief, the odds of target cell-state proportions were divided by the odds of the other cell-state proportions. Odds of cell proportions were calculated as the ratio of cell proportion in the spots of a structure of interest to that in the other spots. Statistical significance was obtained by chi-square analysis (scipy.stats.chi2_contingency) and the P value was corrected with the Benjamini–Hochberg method.
Spatial co-occurrence analysis of cell types
We quantified the degree of co-occurrence between cell types on the basis of their proportions inferred by cell2location. Specifically, since each cell type had an estimated abundance distribution across spatial spots, we calculated the Pearson correlation coefficient for any two given cell types as their co-occurrence rate. This calculation was conducted for each sample separately. Next, the log2 ratio between four Hh-only (control) samples and four Hh/anti-IL10R samples was defined as their fold change in co-occurrence rate and the significance (that is, P value) was assessed by a two-sided Student’s t-test.
Spatial ligand–receptor analysis
Ligand–receptor analysis on Visium data was performed using the Cell2location cell-type abundances and the stLearn package61 (https://github.com/BiomedicalMachineLearning/stLearn). In short, the connectomeDB2020_lit database for mouse was used as a reference of candidate ligand–receptor pairs. The st.tl.cci.run function was used to calculate the significant spots of ligand–receptor interactions within spot mode (distance = None), filtering out any ligand–receptor pairs with no scores for less than 20 spots, and using 10,000 random pairs (n_pairs). P values were corrected with the st.tl.cci.adj_pvals function using false discovery rate, Benjamini–Hochberg (adj_method = ‘fdr_bh’) adjusting by the number of spots tested per ligand–receptor pair (correct_axis = ‘spot’). Spot P values were displayed for particular ligand–receptor pairs (‘Ccl8_Ccr2’ and ‘Cxcl9_Cxcr3’) in the spatial context using the function st.pl.lr_result_plot.
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad). t-Tests were used to compare two unpaired samples. For more than two groups, the ordinary one-way ANOVA was used. No samples were excluded from analysis. Mean with standard deviation shown unless otherwise indicated. Differences were considered statistically significant when P ≤ 0.05. Significance is indicated as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001. Technical replicates were processed and analyses on the same day. Biological replicates are from independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequencing data for scRNA-seq, scTCR-seq and Visium data are available at the European Nucleotide Archive under accession PRJEB57700. Processed data for scRNA-seq and Visium experiments are available for browsing gene expression and downloading at https://treg-gut-niches.cellgeni.sanger.ac.uk/. Source data are provided with this paper.
Code availability
Code scripts and notebooks for analysis in the manuscript are available at https://github.com/Teichlab/treg-gut-niches/.
References
Barnes, M. J. & Powrie, F. Regulatory T cells reinforce intestinal homeostasis. Immunity 31, 401–411 (2009).
Maloy, K. J. & Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504.e497 (2019).
Whibley, N., Tucci, A. & Powrie, F. Regulatory T cell adaptation in the intestine and skin. Nat. Immunol. 20, 386–396 (2019).
Tas, J. M. et al. Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016).
Victora, G. D. et al. Germinal center dynamics revealed by multiphoton microscopy using a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010).
Medaglia, C. et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358, 1622–1626 (2017).
Kullberg, M. C. et al. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J. Exp. Med. 196, 505–515 (2002).
Hue, S. et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 203, 2473–2483 (2006).
Kullberg, M. C. et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 203, 2485–2494 (2006).
Glocker, E. O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Akagbosu, B. et al. Novel antigen-presenting cell imparts Treg-dependent tolerance to gut microbiota. Nature 610, 752–760 (2022).
Lyu, M. et al. ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature 610, 744–751 (2022).
Kedmi, R. et al. A RORγt+ cell instructs gut microbiota-specific Treg cell differentiation. Nature 610, 737–743 (2022).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Pabst, O. et al. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J. Immunol. 177, 6824–6832 (2006).
Pabst, O. et al. Cryptopatches and isolated lymphoid follicles: dynamic lymphoid tissues dispensable for the generation of intraepithelial lymphocytes. Eur. J. Immunol. 35, 98–107 (2005).
Levine, A. G., Arvey, A., Jin, W. & Rudensky, A. Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Kamanaka, M. et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 25, 941–952 (2006).
De Togni, P. et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science 264, 703–707 (1994).
Veiga-Fernandes, H. et al. Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature 446, 547–551 (2007).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Fenton, T. M. et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity 52, 557–570.e6 (2020).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e720 (2020).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Drokhlyansky, E. et al. The human and mouse enteric nervous system at single-cell resolution. Cell 182, 1606–1622.e1623 (2020).
Almanzar, N. et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Corbin, A. L. et al. IRF5 guides monocytes toward an inflammatory CD11c+ macrophage phenotype and promotes intestinal inflammation. Sci. Immunol. 5, eaax6085 (2020).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320.e1322 (2018).
Grenov, A., Hezroni, H., Lasman, L., Hanna, J. H. & Shulman, Z. YTHDF2 suppresses the plasmablast genetic program and promotes germinal center formation. Cell Rep. 39, 110778 (2022).
Xing, S. et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 216, 847–866 (2019).
Yang, B. H. et al. TCF1 and LEF1 control Treg competitive survival and Tfr development to prevent autoimmune diseases. Cell Rep. 27, 3629–3645.e3626 (2019).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Burzyn, D., Benoist, C. & Mathis, D. Regulatory T cells in nonlymphoid tissues. Nat. Immunol. 14, 1007–1013 (2013).
Russler-Germain, E. V., Rengarajan, S. & Hsieh, C. S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 10, 1375–1386 (2017).
Shouval, D. S. et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Do, J. et al. Treg-specific IL-27Rα deletion uncovers a key role for IL-27 in Treg function to control autoimmunity. Proc. Natl Acad. Sci. USA 114, 10190–10195 (2017).
Do, J. S. et al. An IL-27/Lag3 axis enhances Foxp3+ regulatory T cell-suppressive function and therapeutic efficacy. Mucosal Immunol. 9, 137–145 (2016).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Parigi, S. M. et al. The spatial transcriptomic landscape of the healing mouse intestine following damage. Nat. Commun. 13, 828 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Pearson, C. et al. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. eLife 5, e10066 (2016).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Looney, M. R. et al. Stabilized imaging of immune surveillance in the mouse lung. Nat. Methods 8, 91–96 (2011).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151 (2020).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e289 (2019).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Sturm, G. et al. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics 36, 4817–4818 (2020).
Brochet, X., Lefranc, M. P. & Giudicelli, V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508 (2008).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 53, 1698–1711 (2021).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Suo, C. et al. Mapping the developing human immune system across organs. Science 376, eabo0510 (2022).
Gayoso, A. et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40, 163–166 (2022).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Pham, D. et al. Robust mapping of spatiotemporal trajectories and cell–cell interactions in healthy and diseased tissues. Nat. Commun. 14, 7739 (2023).
Acknowledgements
The authors thank the Kennedy Institute of Rheumatology (KIR) Flow Cytometry Facility and the manager, J. Webber, for help with flow cytometry and FACS; the KIR Biomedical Services Unit, especially L. Barker for help with animal care and husbandry; the KIR microscopy facility and manager C. Lagerholm; I. Parisi, B. Stott and R. Cook for tissue processing and staining; A. Lee and M. Attar (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of sequencing data; S. van Dongen and P. V. Mazin and the Teichmann laboratory for discussion and support with scripts. We acknowledge the generous support of the Kennedy Trust for Rheumatology Research, IDRM and Carl Zeiss GMBH for the microscopy facilities (Zeiss 980) used in this research. We acknowledge the generous support of the Kennedy Trust for Rheumatology Research and a Wellcome Trust Multi-User Equipment Grant 202911/Z/16/Z for the microscope purchase (Zeiss 880 multiphoton) and facilities used in this research. Experimental design and summary diagrams were created with BioRender.com. Y.G. was funded by a Wellcome Trust Clinical Research Fellowship (CRTF), grant reference 201224/Z/16/Z. RB-C Grant 315307, Forskerprosjekt 2020, Researcher Project/International Mobility Grant from the Research Council of Norway and travel grant from the Per Brandtzæg’s Fund for Research in Mucosal Immunology. E.E.T. was supported by Wellcome Trust (095688/Z/11/Z and 212240/Z/18/Z, awarded to F.P.), Nuffield Department of Medicine, and MRC core grant reference MC_UU_00008. F.P. was supported by Wellcome Trust (095688/Z/11/Z and 212240/Z/18/Z). This research was funded in whole, or in part, by the Wellcome Trust 212240/Z/18/Z. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
Y.G. designed, performed and analysed the experiments and prepared the manuscript. R.B.-C. conceived and performed the single-cell sequencing analysis, designed, performed and analysed the spatial transcriptomics experiment, and prepared the manuscript. C.X. created the analysis model and assisted with manuscript preparation. A.B. advised on experimental methods, performed immunofluorescence experiments, and assisted with revisions. A.J. and C.H. performed performed immunofluorescence experiments. C.F.P. facilitated and supervised experimentation and provided intellectual input. S.A.T. supervised analysis and manuscript preparation. E.E.T. designed, performed and analysed experiments, supervised the project and prepared the manuscript. F.P. designed experiments and supervised the project and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
C.H. is a current employee of F. Hoffmann-La Roche. S.A.T. is a remunerated member of the scientific advisory board of Element Biosciences, Foresite Labs and Qiagen. S.A.T. is co-founder and equity holder of Transition Bio and EnsoCell, as well as a part-time employee of GlaxoSmithKline. F.P. receives consultancy fees or research support from Janssen, GSK, Genentech, and T-Cypher. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Daniel Mucida and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Immunofluorescent characterisation of intestinal lymphoid structures.
Immunofluorescent expression of DAPI, B220, CD4 and MHCII in the a. caecal patch (scale bar 400 mm). b. organised lymphoid structure (scale bar 200 mm). c. lymphoid aggregate (scale bar 50 mm). d. Gating strategy of FACS sorting of naive T cells from TCRHh donor mice. a-c: Representative images from 2 individual mice over two independent imaging experiments.
Extended Data Fig. 2 TCRHh on a Rag1−/− background validates specificity of TCRHh to Hh.
a. Frequency of Ki-67+ of TCRHh in the tissue (left), MLN (centre left), CP (centre right) and OLS (right) from day 1 to day 14 post cell transfer. b. Co-transfer of naive TCRHhRag1+/− and TCRHhRag1−/− into Hh-colonised hosts. c. Representative FACS plots of TCRHhRag1+/− and TCRHhRag1−/− at the indicated sites. d. Frequency of TCRHhRag1+/− and TCRHhRag1−/− of total CD4 T cells at indicated sites. e. Representative FACS plots (left) and frequency (right) of TCRHhRag1+/− and TCRHhRag1−/− Tregs of TCRHh CD4 T cells. f. Representative FACS plots of TCRHhRag1+/− and TCRHhRag1−/− in Hh-colonised and un-colonised hosts. a: 4 individual mice representative of 2 independent experiments. c-f: 7 individual mice representative of 2 independent experiments. d-e: two-way ANOVA using a Šidák’s multiple comparisons test with individual variances computed for each comparison.
Extended Data Fig. 3 FTY720 treatment supports tissue as a dominant site of TCR signalling and IL-10 production.
a. Geometric MFI (left) and frequency (right) of Nur77GFP positive of TCRHh Tregs at the indicated sites. b. Frequency of IL-10+ of TCRHh Tregs at the indicated sites. c. Histogram overlay (left) and quantification of geometric MFI (right) of TCRHhNur77GFP co-cultured with Hh-pulsed BMDCs at the indicated time points. d. Transfer of TCRHhNur77GFP into FTY720-treated host mice. e. Representative FACS plots (left) and quantification (right) of TCRβ of total CD45 in peripheral blood. f. Geometric MFI of total TCRHhNur77GFP in FTY720 treated hosts at the indicated sites. g. Geometric MFI of total TCRHhNur77GFP in the tissue of control and FTY720 treated hosts. h. Frequency of IL-10GFP positive among total TCRHh in FTY720 treated hosts. i. Frequency of IL-10GFP positive among total TCRHh in the tissue of control and FTY720 treated hosts. j. Absolute TCRHh cell numbers in tissue of control and FTY720-treated hosts. a: 6 individual mice representative of 2 independent experiments. b: 5 individual mice representative of 2 independent experiments. c: 3 technical replicates representative of 2 independent experiments. e-j: 4 WT and 5 FTY720 treated mice representative of 2 independent experiments. a-b, f, h: one-way ANOVA using a Tukey’s multiple comparisons test. e, g, i-j: two-tailed t test.
Extended Data Fig. 4 IL-10 is critical for maintaining homeostasis but LNs are not required for the pathogenic response to Hh in LTa−/− mice.
a. Representative images of the caecal patch (left) and OLS (right) in Hh-colonised WT and LTa−/− hosts. b. Representative FACS plots of TCRHh in tissue at 10 days post naïve TCRHh cell transfer. c. Absolute TCRHh cell count in tissue of Hh-colonised WT and LTa−/− hosts. d. Absolute host CD4 + T cell count in tissue in Hh-colonised WT and LTa−/− hosts. e. Representative FACS plots (left) and frequency of Tregs among TCRHh (right) in tissue of WT and LTa−/− hosts at 10 days post naïve TCRHh transfer. f. Representative FACS plots (left) and frequency of IL-10+ among TCRHh (right) in tissue of WT and LTa−/− hosts at 10 days post naïve TCRHh transfer. g. Absolute cell count of IL-10+ among TCRHh in tissue of WT and LTa−/− hosts at 10 days post naïve TCRHh transfer. h. Frequency of IL-10+ of TCRHh Tregs in tissue of Hh-colonised WT and LTa−/− hosts at 10 days post naïve TCRHh transfer. i. Colon histopathology score of Hh-colonised WT and LTa−/− mice with and without anti-IL-10R treatment. j. Two photon in vivo live imaging of transferred naive TCRHhCD2dsRedNur77GFP or TCRHhCD2dsRedIL-10GFP into Hh-colonised Foxp3GFP hosts. The caecum LA and LP were imaged. a: Representative images from 3 individual mice across 2 independent experiments. b-h: 4 WT and 4 LTa−/− individual mice representative of 2 independent experiments. i: 5 WT and 5 LTa−/− individual mice representative of 2 independent experiments. c-h: two-tailed t test. i: one-way ANOVA using a Tukey’s multiple comparisons test.
Extended Data Fig. 5 FACS cell sorting of photoactivated cells and tissue memory heterogeneity.
a. Representative images of pre- and post-photoactivation of LA (top) and T cell zone of CP (bottom). b. Gating strategy of cell sorting by FACS of GFP+ cells in the caecal patch (top left), MLN (bottom left) and caecum LA and LP (right). GFP gate set on un-photoactivated samples. c. UMAP visualisation of total cell subsets (left) with detailed cell type annotations (right). d. Bar chart showing total cell subsets across all tissue niches (top left) and cell numbers (bottom left). UMAP visualisation of cell lineages by gut region (bottom right). e. UMAP of CD4 memory, Th17, and gd T cells subsets (top). Overlap of cell location (bottom). f. Gene overlays of CD4 memory subsets from all tissues. g. Gene expression of canonical Th1 and Th17 genes expressed by the CD4 memory subsets. h. Frequency bar chart showing the subsets of CD4 T cells in the LA and LP. a-h: Representative of 6 combined mice.
Extended Data Fig. 6 Developmental relationships between transcriptionally distinct cTregs, eTregs and proliferating Tregs.
a. Heatmap of differentially expressed genes in cTreg, eTreg and proliferating Tregs. b. UMAP of Treg subsets per location, filtering out the subsets with less than 10 cells in a given location (i.e. excluding eTregs_MLN, eTregs_CP, cTregs_LA, cTregs_LP and Prolif-Tregs_CP). Arrows depict summarised scVelo differentiation trajectories (top). Heat map overlay on the Treg UMAP indicating velocity pseudotime, as calculated by scVelo (bottom). c. Velocity analysis of genes that vary across the differentiation trajectory of all Treg subsets. d. Rorc and Gata3 expression of Tregs with Maf expression overlaid in red. e. Heatmap of differentially expressed genes between eTregs in MLN and LP. f. UMAP of the total Treg population with distributions of Treg subsets across locations (top) and overlay of TCRHh Tregs (below). g. UMAP overlay of clone size (top) and clone specificity (bottom) of TCRHh within total T cell populations. h. Barchart showing the distribution of most expanded TCRHh and host clones by location (top) and T cell subset (bottom). i. Heatmap of differentially expressed genes of host LP vs LA eTregs. Blue lines indicate gene expression profile of TCRHh eTregs in LP and LA. a-i: Representative of 6 combined mice.
Extended Data Fig. 7 Treatment with FTY720 does not affect the phenotype and proliferation of TCRHh blocked with anti-IL-10R.
a. TCRHh were transferred into Hh colonised host mice, treated with daily FTY720 from day 15 and anti-IL-10R at day 17. b. Frequency of Foxp3 among total TCRHh CD4 T cells (left), geometric MFI of Foxp3 expression in TCRHh Tregs (centre) and geometric MFI of c-MAF expression in TCRHh Tregs (right) in control and anti-IL-10R treated mice at day 20. c. Ki-67 of total CD4 T cells of TCRHh and host cells (left), Ki-67 of TCRHh Tregs (centre) and Ki-67 of TCRHh Th17 cells (right) in control and anti-IL-10R treated mice at day 24 with and without FTY720 blockade. d. Representative FACS plot of c-MAF versus Ki-67 in TCRHh Tregs (left) and c-MAF geometric MFI of Ki-67+ vs Ki-67- TCRHh Tregs (right). e. Geometric MFI of Foxp3 (left) and c-MAF (right) of TCRHh Tregs in control and anti-IL-10R treated mice with and without FTY720 blockade. g. Expression of Ifnγ (left), Il17α (centre) and Il6 (right) in caecum tissue by qPCR in control and anti-IL-10R treated mice at day 24. g. Expression of Il27 (left), Il10 (centre left), Il2 (centre right) and Il23a (right) in caecum tissue by qPCR in control and anti-IL-10R treated mice at day 24. h. Representative H and E staining (left top and bottom) and histopathology score (right top) of caecum tissue from control and anti-IL-10R treated mice at day 24. Scale bar represents 1 mm. i. Colon histopathology score in control and anti-IL-10R treated mice with and without FTY720 blockade. j. Representative immunofluorescent staining of Foxp3 and Ki-67 in LA (left, arrows mark Ki-67 negative Tregs), control LP (centre, arrows mark Ki-67 negative Tregs), and anti-IL-10R treated LP (right, arrows mark Ki-67 positive Tregs). k. Frequency of Ki-67 positive Tregs in the LP of control and anti-IL-10R treated mice (left) and frequency of Ki-67 positive Tregs in the LP and LA in both control and anti-IL-10R treated mice (right). b-i: 6 individual mice per group representative of 2 independent experiments. f: 11 control and 11 anti-IL-10R treated mice combined over 2 independent experiments. J-k: 5 individual mice per group representative of 2 independent experiments. b, d, f, g, k: two-tailed t test. c, e, i: one-way ANOVA using a Tukey’s multiple comparisons test.
Extended Data Fig. 8 Validation of micro-niches by ST analysis and NICHE-seq.
a. Representative H&E image with manually annotated LA and LP from Hh/anti-IL-10R. b. Visual representation of cell cycle scoring of Hh (top) and Hh/anti-IL-10R (middle) treated gut swiss-rolls with quantification of S score (bottom left) and G2M score (bottom). c. Cell type decomposition of gut swiss rolls in Hh (left) and Hh/anti-IL-10R (right) treated mice. Dotplots showing NMF weights of cell types (columns) across NMF factors (rows), which correspond to tissue microenvironments (normalized across factors per cell type by dividing by maximum values). d. Identification of microglia/enteric neurone associated factor 5 (left) and LA associated factor 3 (right). e. Hh/anti-IL-10R swiss roll H&E. Localisation of LA associated factor 3 by NMF -based decomposition and its overlying RNA capture spots (top left). Localisation of LA and its overlying capture spot as defined by manual annotation (top centre and right). Localisation and normalized cell type abundance of eMBCs (bottom left), naïve CD8 + T cells (bottom centre) and LTi-like ILC3s (bottom right) in representative LA. f. Dotplot of Il10 and Il6 gene expression in the LA and LP by ST analysis (left). Violin plot of relative distribution of Il10 gene expression in the LA and LP by ST analysis (right). g. Cell type abundance in manually annotated LA and LP regions of Hh (top) and Hh/anti-IL-10R (bottom). Relative weights, normalized across LA/LP for every cell type. Cell type loadings represented by dot size and colour. h. Cell state enrichment (odds ratio) in the manually annotated LA vs the rest of the tissue in Hh (top) and Hh/anti-IL-10R (bottom). Statistically significant enrichments (chi-square test, p < 0.05) are shown in magenta-pink. Data show value ± SD. i. Cell state enrichment (odds ratio) in the manually annotated LA vs the rest of the tissue with muscularis mucosa removed in Hh (top) and Hh/anti-IL-10R (bottom). Statistically significant enrichments (chi-square test, p < 0.05) are shown in magenta-pink. Data show value ± SD. a-i: Representative of 4 individual mice per group (Hh and Hh/anti-IL-10R sequenced from two Visium slides.
Extended Data Fig. 9 Expression of cytokines and chemokine for eTreg function.
a. Co-occurrence analysis of Hh (left top) and Hh/anti-IL-10R (left bottom) with the ratio of the two conditions (right). Size of dots represents p-values and the colour represents fold change. Number of spots analysed after removing Muscularis-associated NMF factors (4405 spots) are Hh/anti-IL-10R: 9572 spots and Hh: 8815 spots. b. Chord diagrams showing significant interactions of Tregs with lymphoid and myeloid cells in LP (top) and LA (bottom). Restricted to cell types with >30 cells per region and significance p < 0.05 for interactions between any cell pair. c.Dot plot showing relative expression by NICHE-seq analysis of survival cytokine genes expressed by all the cell types with >30 cells in LA or LP regions (left) and their receptors on Treg populations (right). b. NICHE-seq analysis UMAP visualisation of T-cell subsets (left) with overlay of Ccr5 and Ccr2 expression (right). c. Dot plot showing expression of Ccr2 and Ccr5 and their ligands (Ccl7 and Ccl8) in myeloid cells, T cells and ILCs isolated from the LA and LP NICHE-seq analysis. d. Localisation of Ccr2 (top left), Cxcr3 (top centre) and Cxcr4 (bottom left) expression in LA and LP by ST analysis on H&E (bottom centre). Violin plots of relative distribution of Ccr2, Cxcr3 and Cxcr4 expression (top right). Dot plot of Ccr2, Cxcr3 and Cxcr4 expression in the LA and LP by ST analysis (bottom right). e. Ligand-receptor analysis on Visium data. Adjusted p-values were calculated per spot using the package stLearn and displayed in the spatial context for particular ligand-receptor pairs (‘Ccl8_Ccr2’ and ‘Cxcl9_Cxcr3’) in two representative Hh samples (above, Hh mouse 2 and below Hh mouse 4). a-c: One sequencing run from 6 combined mice. d-e: Two representative samples of 4 individual mice per group (Hh and Hh/anti-IL-10R sequenced from two Visium slides.
Supplementary information
Supplementary Figs. 1–5
Detailed information regarding single-cell bioinformatics analysis workflow, cell type determination, and spatial transcriptomics analysis.
Supplementary Table 1
Differentially expressed genes for each cluster, grouped by cell lineages. Statistical test used was Wilcoxon rank-sum implemented in Scanpy v.1.7. P value correction was performed using the Benjamini–Hochberg method.
Supplementary Table 2
Compiled gene lists used for the different scores.
Supplementary Video 1
Rotation of a 3D volume of caecum imaged with a two-photon microscope shows a LA highlighted by green Treg cells with TCRHhCD2DsRedNur77GFP cells with red cytoplasm and green nuclei.
Supplementary Video 2
Time lapse of a TCRHhCD2DsRedNur77GFP cell in the caecum LP demonstrating motility through the tissue.
Supplementary Video 3
Time lapse of TCRHhCD2DsRedIl10GFP T cells in the caecum LP demonstrating motility through the tissue.
Supplementary Video 4
Time lapse of IL-10+ TCRHh cell interacting with CD206+ macrophages in the caecum LP.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gu, Y., Bartolomé-Casado, R., Xu, C. et al. Immune microniches shape intestinal Treg function. Nature 628, 854–862 (2024). https://doi.org/10.1038/s41586-024-07251-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-07251-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.