Abstract
The accumulation of metabolic waste is a leading cause of numerous neurological disorders, yet we still have only limited knowledge of how the brain performs self-cleansing. Here we demonstrate that neural networks synchronize individual action potentials to create large-amplitude, rhythmic and self-perpetuating ionic waves in the interstitial fluid of the brain. These waves are a plausible mechanism to explain the correlated potentiation of the glymphatic flow1,2 through the brain parenchyma. Chemogenetic flattening of these high-energy ionic waves largely impeded cerebrospinal fluid infiltration into and clearance of molecules from the brain parenchyma. Notably, synthesized waves generated through transcranial optogenetic stimulation substantially potentiated cerebrospinal fluid-to-interstitial fluid perfusion. Our study demonstrates that neurons serve as master organizers for brain clearance. This fundamental principle introduces a new theoretical framework for the functioning of macroscopic brain waves.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data necessary for the conclusions of the study are available in the main text, figures and extended data. Representative raw widefield and MRI images are available from Zenodo (https://doi.org/10.5281/zenodo.10440376). Source data are provided with this paper.
Code availability
The Matlab codes used in this study are available from Zenodo (https://doi.org/10.5281/zenodo.10440376).
References
Iliff, J. J. et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4, 147ra111 (2012).
Xie, L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013).
Magistretti, P. J. & Allaman, I. Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 19, 235–249 (2018).
Laughlin, S. B., de Ruyter van Steveninck, R. R. & Anderson, J. C. The metabolic cost of neural information. Nat. Neurosci. 1, 36–41 (1998).
Kaplan, L., Chow, B. W. & Gu, C. Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nat. Rev. Neurosci. 21, 416–432 (2020).
Oliver, G., Kipnis, J., Randolph, G. J. & Harvey, N. L. The lymphatic vasculature in the 21st century: novel functional roles in homeostasis and disease. Cell 182, 270–296 (2020).
Hablitz, L. M. & Nedergaard, M. The glymphatic system. Curr. Biol. 31, R1371–R1375 (2021).
Rasmussen, M. K., Mestre, H. & Nedergaard, M. Fluid transport in the brain. Physiol. Rev. 102, 1025–1151 (2022).
Lei, Y., Han, H., Yuan, F., Javeed, A. & Zhao, Y. The brain interstitial system: anatomy, modeling, in vivo measurement, and applications. Prog. Neurobiol. 157, 230–246 (2017).
Fultz, N. E. et al. Coupled electrophysiological, hemodynamic, and cerebrospinal fluid oscillations in human sleep. Science 366, 628–631 (2019).
Hablitz, L. M. et al. Increased glymphatic influx is correlated with high EEG delta power and low heart rate in mice under anesthesia. Sci. Adv. 5, eaav5447 (2019).
Jiang-Xie, L. F. et al. A common neuroendocrine substrate for diverse general anesthetics and sleep. Neuron 102, 1053–1065.e4 (2019).
Buzsaki, G. Large-scale recording of neuronal ensembles. Nat. Neurosci. 7, 446–451 (2004).
Buzsaki, G., Anastassiou, C. A. & Koch, C. The origin of extracellular fields and currents—EEG, ECoG, LFP and spikes. Nat. Rev. Neurosci. 13, 407–420 (2012).
Scammell, T. E., Arrigoni, E. & Lipton, J. O. Neural circuitry of wakefulness and sleep. Neuron 93, 747–765 (2017).
Buzsaki, G. & Draguhn, A. Neuronal oscillations in cortical networks. Science 304, 1926–1929 (2004).
Weber, F. & Dan, Y. Circuit-based interrogation of sleep control. Nature 538, 51–59 (2016).
Akeju, O. & Brown, E. N. Neural oscillations demonstrate that general anesthesia and sedative states are neurophysiologically distinct from sleep. Curr. Opin. Neurobiol. 44, 178–185 (2017).
Chauvette, S., Crochet, S., Volgushev, M. & Timofeev, I. Properties of slow oscillation during slow-wave sleep and anesthesia in cats. J. Neurosci. 31, 14998–15008 (2011).
Steinmetz, N. A. et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science https://doi.org/10.1126/science.abf4588 (2021).
Destexhe, A., Contreras, D. & Steriade, M. Spatiotemporal analysis of local field potentials and unit discharges in cat cerebral cortex during natural wake and sleep states. J. Neurosci. 19, 4595–4608 (1999).
Buzsáki, G. Rhythms of the Brain (Oxford Univ. Press, 2006).
Magnus, C. J. et al. Ultrapotent chemogenetics for research and potential clinical applications. Science https://doi.org/10.1126/science.aav5282 (2019).
Luo, L. Principles of Neurobiology 2nd edn (Garland Science, 2020).
Turecek, J., Lehnert, B. P. & Ginty, D. D. The encoding of touch by somatotopically aligned dorsal column subdivisions. Nature 612, 310–315 (2022).
Eide, P. K., Vinje, V., Pripp, A. H., Mardal, K. A. & Ringstad, G. Sleep deprivation impairs molecular clearance from the human brain. Brain 144, 863–874 (2021).
Miyawaki, H. & Diba, K. Regulation of hippocampal firing by network oscillations during sleep. Curr. Biol. 26, 893–902 (2016).
Iliff, J. J. et al. Brain-wide pathway for waste clearance captured by contrast-enhanced MRI. J. Clin. Invest. 123, 1299–1309 (2013).
Chen, R. et al. Deep brain optogenetics without intracranial surgery. Nat. Biotechnol. 39, 161–164 (2021).
Marshel, J. H. et al. Cortical layer-specific critical dynamics triggering perception. Science https://doi.org/10.1126/science.aaw5202 (2019).
Anikeeva, P. et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat. Neurosci. 15, 163–170 (2011).
Harris, K. D. & Thiele, A. Cortical state and attention. Nat. Rev. Neurosci. 12, 509–523 (2011).
Smith, A. J. & Verkman, A. S. The “glymphatic” mechanism for solute clearance in Alzheimer’s disease: game changer or unproven speculation? FASEB J. 32, 543–551 (2018).
Hladky, S. B. & Barrand, M. A. The glymphatic hypothesis: the theory and the evidence. Fluids Barriers CNS 19, 9 (2022).
Pulido, R. S. et al. Neuronal activity regulates blood–brain barrier efflux transport through endothelial circadian genes. Neuron 108, 937–952.e7 (2020).
Holstein-Ronsbo, S. et al. Glymphatic influx and clearance are accelerated by neurovascular coupling. Nat. Neurosci. 26, 1042–1053 (2023).
Massimini, M., Huber, R., Ferrarelli, F., Hill, S. & Tononi, G. The sleep slow oscillation as a traveling wave. J. Neurosci. 24, 6862–6870 (2004).
Lubenov, E. V. & Siapas, A. G. Hippocampal theta oscillations are travelling waves. Nature 459, 534–539 (2009).
Zhang, H. & Jacobs, J. Traveling theta waves in the human hippocampus. J. Neurosci. 35, 12477–12487 (2015).
Mestre, H. et al. Aquaporin-4-dependent glymphatic solute transport in the rodent brain. eLife https://doi.org/10.7554/eLife.40070 (2018).
Smith, A. J., Yao, X., Dix, J. A., Jin, B. J. & Verkman, A. S. Test of the ‘glymphatic’ hypothesis demonstrates diffusive and aquaporin-4-independent solute transport in rodent brain parenchyma. eLife https://doi.org/10.7554/eLife.27679 (2017).
Pla, V. et al. A real-time in vivo clearance assay for quantification of glymphatic efflux. Cell Rep. 40, 111320 (2022).
Tononi, G. & Edelman, G. M. Consciousness and complexity. Science 282, 1846–1851 (1998).
Shein-Idelson, M., Ondracek, J. M., Liaw, H. P., Reiter, S. & Laurent, G. Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 352, 590–595 (2016).
Yap, M. H. W. et al. Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat. Commun. 8, 1815 (2017).
Leung, L. C. et al. Neural signatures of sleep in zebrafish. Nature 571, 198–204 (2019).
Iaccarino, H. F. et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235 (2016).
Martorell, A. J. et al. Multi-sensory gamma stimulation ameliorates Alzheimer’s-associated pathology and improves cognition. Cell 177, 256–271.e22 (2019).
Luo, L. Architectures of neuronal circuits. Science 373, eabg7285 (2021).
Drieu, A. et al. Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid. Nature 611, 585–593 (2022).
Franklin, K. B. J. & Paxinos, G. The mouse brain in stereotaxic coordinates 4th edn (Academic Press, 2013).
Ayloo, S. et al. Pericyte-to-endothelial cell signaling via vitronectin-integrin regulates blood–CNS barrier. Neuron 110, 1641–1655.e6 (2022).
Araragi, N., Alenina, N. & Bader, M. Carbon-mixed dental cement for fixing fiber optic ferrules prevents visually triggered locomotive enhancement in mice upon optogenetic stimulation. Heliyon 8, e08692 (2022).
Kass, R. E., Eden, U. T. & Brown, E. N. Analysis of Neural Data (Springer, 2014).
Kramer, M. A. & Eden, U. T. Case studies in neural data analysis: a guide for the practicing neuroscientist (The MIT Press, 2016).
Cohen, M. X. Analyzing neural time series data: theory and practice (The MIT Press, 2014).
Rossant, C. et al. Spike sorting for large, dense electrode arrays. Nat. Neurosci. 19, 634–641 (2016).
Prerau, M. J., Brown, R. E., Bianchi, M. T., Ellenbogen, J. M. & Purdon, P. L. Sleep neurophysiological dynamics through the lens of multitaper spectral analysis. Physiology 32, 60–92 (2017).
Acknowledgements
We thank S. Smith for editing the manuscript; A. Impagliazzo for her help on the illustration for Extended Data Fig. 9d; E. Griffin and A. Apaw for handling the mouse colony; S. Brophy for laboratory management; J. Quirk for his guidance on the acquisition and analysis of MRI images; and members of the Kipnis Laboratory for comments and suggestions. This study is supported by National Institutes of Health (NIH) DP1AT010416 (to J.K.), R01AT011419 (to J.K.), Barnes–Jewish Hospital investigators program (to J.K.) from Washington University School of Medicine, and a gift from the Neuroscience Innovation Foundation.
Author information
Authors and Affiliations
Contributions
L.-F.J.-X. conceived the original idea, designed and performed most of the experiments and analysed the data. A.D. helped design, execute and analyse MRI images. K.B. helped with stereotaxic surgeries, brain sectioning using the cryostat microtome, immunofluorescence staining, widefield imaging and data analysis. D.Q. helped with cryostat sectioning and widefield imaging. I.S. helped with surgical procedures, maintaining the mouse colony and post-surgery care of animals. J.K. participated in experimental design, provided intellectual guidance, resources and supervised the entire study. L.-F.J.-X. and J.K. wrote the paper with the input from all authors.
Corresponding authors
Ethics declarations
Competing interests
J.K. is a co-founder of Rho Bio. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Axel Montagne and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of EEGs and ionic waves in ISF of hippocampus during wake and ketamine anesthesia with chemogenetic manipulation.
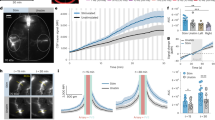
a, Schematic of electrode location in PSAM-expressed hippocampus. bi, Power spectra of frontal EEG (fEEG) and parietal EEG (pEEG) during wake, ketamine anesthesia (Ket) and Ket + uPSEM792. n = 5 mice for wake; n = 4 mice for Ket and Ket + uPSEM792. Shaded areas denote 95 percent confidence intervals for the mean. bii, Power spectra of ISF waves in hippocampus during wake, ketamine anesthesia with or without chemogenetic inhibition. n = 136 channes from 5 mice. Shaded areas denote 95 percent confidence intervals for the mean. c-e, Cross-frequency coupling analysis in hippocampal ISF during ketamine anesthesia, scale bar: 300 ms and 200 µV. Raw and filter traces (c), phase and power extraction (d), and polar plot across all 28-recording channels (e).
Extended Data Fig. 2 Analysis of neuronal spikes in hippocampus during wake and ketamine anesthesia with chemogenetic perturbation.
a, Examples of spike waveforms. b, Spike rate for isolated units during wakefulness and ketamine (n = 34 units from 5 mice). Left, statistical summary of unit spike rates; right, normalized activity heatmap. Two-sided Wilcoxon signed-rank test. p < 0.0001. c, Instant amplitude of field potential coupled with neuronal spikes (n = 34 units from 5 mice). Two-sided Wilcoxon signed-rank test. p < 0.0001. d, Spike rate for isolated units during wakefulness and ketamine with chemogenetic inhibition (n = 35 units from 5 mice). Left, statistical summary of unit spike rates; right, normalized activity heatmap. Two-sided Wilcoxon signed-rank test. p < 0.0001. *** p < 0.001.
Extended Data Fig. 3 Molecular infiltration analysis on additional brain regions.
a, Representative images from PSAM-expressing animals. Left column displays composite images (GFP, Dex, and DAPI) from anterior to posterior (top to bottom); while the right column shows corresponding tracer-only (Dex) images. Statistical summary for tracer infiltration in GFP group. (b) and PSAM group (c) across multiple brain regions: Ant DC, anterior dorsal cortex; Ant VC, anterior ventral cortex; Post DC, posterior dorsal cortex; Post VC, posterior ventral cortex; Hypo, hypothalamus. n = 7 mice for both GFP and PSAM groups. Scale bar: 500 µm. Two-sided Paired-t test. n.s., not significant.
Extended Data Fig. 4 Acute neuronal inhibition does not affect GFAP expression and blood-brain barrier integrity.
a, GFAP staining after uPSEM792 treatment (3 mg/kg, i.p.) in GFP group (left panel, n = 4) and PSAM group (right panel, n = 4). GFAP, glial fibrillary acidic protein. Scale bar: 500 µm. Paired-t test. n.s., not significant. b, Blood-brain barrier leakage assay after uPSEM792 treatment (3 mg/kg, i.p.) in GFP group (left panel, n = 4) and PSAM group (right panel, n = 4). Scale bar: 500 µm. Two-sided paired-t test. n.s., not significant.
Extended Data Fig. 5 Characterization of EEGs and ISF waves in hippocampus during natural sleep-wake cycle with chemogenetic inhibition.
a, Representative traces of EEG, EMG, and ionic waves in the hippocampus during wake, NREM, and REM with chemogenetic inhibition. Top right scale bar: 200 ms and 200 µV for EEGs; bottom right scale bar: 200 ms and 200 µV for LFPs. b, Power spectra of cortical EEGs (n = 4 animals) before and after chemogenetic inhibition. c, Power spectra of hippocampal ISF before (n = 111 channels from 4 animals) and after (n = 109 channels from 4 animals) chemogenetic inhibition. Shaded areas denote 95 percent confidence intervals for the mean.
Extended Data Fig. 6 Analysis of neuronal spikes in hippocampus during natural sleep-wake cycle with chemogenetic perturbation, the asymmetry of CSF perfusion between sleep and wake.
a, Examples of spike waveforms. b-c, Spike rate for isolated units (n = 37 units from 4 mice) underlying wake, NREM, and REM with chemogenetic inhibition. b, Statistical summary of unit spike rates; c, Normalized activity heatmap. d, Instant amplitude coupled with neuronal spikes (n = 37 units from 4 mice). Two-sided Wilcoxon signed-rank test with Bonferroni correction. p = 0.0001 (Wake vs NREM) and p < 0.0001 (Wake vs REM) e, Asymmetry of CSF perfusion between wake (n = 10 animals) and sleep (n = 9 animals) conditions measured as percentage of contralateral (Contra) side. Two-sided Student t-test. p = 0.0045. ** p < 0.01, *** p < 0.001.
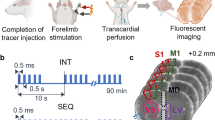
Extended Data Fig. 7 EEG/EMG characterization after acute intracisterna magna (ICM) injection.
a, Representative spectrograms from fEEG (top row), pEEG (second row), EMG (third row), hypnogram (fourth row) after acute ICM injection. b, Representative recording traces in three different brain states, wake, NREM, and REM. Blue trace, fEEG; red trace, pEEG, black trace, EMG. Scale bar: 500 ms and 200 µV. c, Power spectrum analysis across wake, NREM, and REM in the frontal EEG (top) and parietal (bottom) EEG channels. d, Percentage of time spent in Wake, NREM, and REM sleep., n = 3 animals. fEEG, frontal EEG; pEEG, parietal EEG.
Extended Data Fig. 8 MRI, the time window for molecular clearance, and the efficacy and duration of chemogenetic inhibition.
a-b, Paravascular flow is largely preserved during local neuronal inhibition in hippocampus. Left panel, GFP group; right panel, PSAM group. GadoSpinP, large molecular tracer (~200 kDa) used to visualize para-vascular flow. ICM, intracisterna magna injection. n = 3 mice for GFP group; n = 4 mice for PSAM group. Data presented as mean ± s.e.m. c, 3−7 h after ICM (3 kD Dextran-TexasRed, yellow) injection mainly captures molecular clearance phase. n = 6 mice for 3-hour group; n = 8 mice for 7-hour group. Two-sided Mann-Whitney test, p = 0.0426. Scar bar = 500 µm. d, Chemogenetic inhibition lasts for 5 h after uPSEM792 injection across three brain states, wake, NREM, REM. n = 109 channels from 4 animals. Shaded areas denote 95 percent confidence intervals for the mean. *p < 0.05.
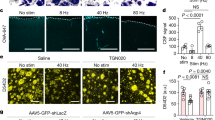
Extended Data Fig. 9 Validation of optogenetic toolkits.
a, Transcranial activation of neurons revealed by Fos staining. Left, representative images; right, statistical summary (n = 4 animals). Contra, contralateral side of the hippocampus. Scale bar: 500 µm. Two-sided paired-t test. p = 0.0031. b, Representative field potential traces with photo-stimulations in ChRmine-expressing animals. Slow stimulation: 1 Hz, 50 ms per TTL pulse; theta stimulation: 8 Hz, 6.25 ms per TTL pulse. Scale bar: 200 ms and 300 µV. c, Quantification of slow wave (0.5−4 Hz) power and theta wave (6−10 Hz) power from optrode recording experiment (n = 54 channels from 2 animals). d, Illustration of the principle that neurons firing together shower together. ** p < 0.01.
Extended Data Fig. 10 Wave phase progression analysis across electrophysiological recording channels in hippocampus during different brain states.
a-c, Representative slow (0.5−4 Hz) filtered traces (a) from hippocampal field potential recordings during ketamine anesthesia, corresponding phases (b) extracted with Hilbert method, and averaged phase shift (c). Scale bar: 200 ms and 200 µV. d-f, Representative slow (0.5−4 Hz) filtered traces (d) from hippocampal field potential recordings during NREM sleep, corresponding phases (e) extracted with Hilbert method, and averaged phase shift (f). Scale bar: 100 ms and 200 µV. g-i, Representative theta (6–10 Hz) filtered traces (g) from hippocampal field potential recordings during REM sleep, corresponding phases (h) extracted with Hilbert method, and averaged phase shift (i). Scale bar: 100 ms and 200 µV.
Supplementary information
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang-Xie, LF., Drieu, A., Bhasiin, K. et al. Neuronal dynamics direct cerebrospinal fluid perfusion and brain clearance. Nature 627, 157–164 (2024). https://doi.org/10.1038/s41586-024-07108-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-07108-6
This article is cited by
-
Synchronized neuronal activity drives waste fluid flow
Nature (2024)
-
Neuronal activity drives glymphatic waste clearance
Nature Reviews Neurology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.