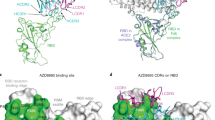
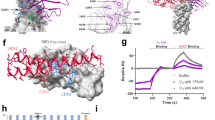
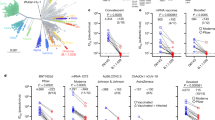
Abstract
A severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariant, BA.2.86, has emerged and spread to numerous countries worldwide, raising alarm because its spike protein contains 34 additional mutations compared with its BA.2 predecessor1. We examined its antigenicity using human sera and monoclonal antibodies (mAbs). Reassuringly, BA.2.86 was no more resistant to human sera than the currently dominant XBB.1.5 and EG.5.1, indicating that the new subvariant would not have a growth advantage in this regard. Importantly, sera from people who had XBB breakthrough infection exhibited robust neutralizing activity against all viruses tested, suggesting that upcoming XBB.1.5 monovalent vaccines could confer added protection. Although BA.2.86 showed greater resistance to mAbs to subdomain 1 (SD1) and receptor-binding domain (RBD) class 2 and 3 epitopes, it was more sensitive to mAbs to class 1 and 4/1 epitopes in the ‘inner face’ of the RBD that is exposed only when this domain is in the ‘up’ position. We also identified six new spike mutations that mediate antibody resistance, including E554K that threatens SD1 mAbs in clinical development. The BA.2.86 spike also had a remarkably high receptor affinity. The ultimate trajectory of this new SARS-CoV-2 variant will soon be revealed by continuing surveillance, but its worldwide spread is worrisome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All experimental data are provided in the manuscript. Materials used in this study will be available under an appropriated Materials Transfer Agreement. Antigenic maps were generated using the Racmacs package (v.1.1.4, https://acorg.github.io/Racmacs/) in R v.4.0.3. Severe acute respiratory syndrome coronavirus 2 spike sequences were downloaded from the Global Initiative on Sharing Avian Flu Data database (https://www.gisaid.org/). The structures used for analysis in this study are available from the Protein Data Bank under identification codes 7KRR, 7WKA, 8D8Q, 7MMO, 7TAS, 7TCA and 7ZF7.
References
Khare, S. et al. GISAID’s role in pandemic response. China CDC Wkly 3, 1049–1051 (2021).
World Health Organization. Statement on the fifteenth meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. WHO https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (2023).
Wang, Q. et al. Antibody neutralisation of emerging SARS-CoV-2 subvariants: EG.5.1 and XBC.1.6. Lancet Infect. Dis. 23, e397–e398 (2023).
Wang, Q. et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286 (2023).
Miller, J. et al. Substantial neutralization escape by SARS-CoV-2 Omicron variants BQ.1.1 and XBB.1. N. Engl. J. Med. 388, 662–664 (2023).
Yue, C. et al. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis. 23, 278–280 (2023).
UK Health Security Agency. SARS-CoV-2 variant surveillance and assessment: technical briefing 53. UKHSA https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings/sars-cov-2-variant-surveillance-and-assessment-technical-briefing-53 (2023).
Wang, Q. et al. Evolving antibody evasion and receptor affinity of the Omicron BA.2.75 sublineage of SARS-CoV-2. iScience 26, 108254 (2023).
Wang, Q. et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 603–608 (2022).
Liu, L. et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 375, 449–454 (2022).
Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529 (2023).
Nutalai, R. et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell 185, 2116–2131 (2022).
Cao, Y. et al. Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res. 31, 732–741 (2021).
Zost, S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020).
Wang, K. et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 603, 919–925 (2022).
Zhou, B. et al. A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge. Nat. Commun. 13, 3589 (2022).
Wang, L. et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 373, eabh1766 (2021).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Westendorf, K. et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 39, 110812 (2022).
Liu, C. et al. The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Cell Host Microbe 30, 53–68 (2022).
Cao, Y. et al. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep. 41, 111845 (2022).
Liu, L. et al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci. Transl. Med. 14, eabn6859 (2022).
Wang, Z. et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Immunity 55, 998–1012 (2022).
Hong, Q. et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 604, 546–552 (2022).
Guenthoer, J. et al. Identification of broad, potent antibodies to functionally constrained regions of SARS-CoV-2 spike following a breakthrough infection. Proc. Natl Acad. Sci. USA 120, e2220948120 (2023).
Yuan, M. et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020).
Wang, Q. et al. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe 30, 1512–1517.e4 (2022).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310 (2020).
Qu, P. et al. Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe 30, 1518–1526.e4 (2022).
Han, P. et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 185, 630–640 (2022).
Lasrado, N. et al. Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86. Vaccine 41, 6904–6909 (2023).
An, Y. et al. SARS-CoV-2 Omicron BA.2.86: less neutralization evasion compared to XBB sub-variants. Preprint at bioRxiv https://doi.org/10.1101/2023.09.26.559580 (2023).
Khan, K. et al. Evolution and neutralization escape of the SARS-CoV-2 BA.2.86 subvariant. Preprint at medRxiv https://doi.org/10.1101/2023.09.08.23295250 (2023).
Yang, S. et al. Antigenicity and infectivity characterisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(23)00573-X (2023).
Uriu, K. et al. Transmissibility, infectivity, and immune evasion of the SARS-CoV-2 BA.2.86 variant. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(23)00575-3 (2023).
Sheward, D. J. et al. Sensitivity of the SARS-CoV-2 BA.2.86 variant to prevailing neutralising antibody responses. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(23)00588-1 (2023).
Chalkias, S. et al. Safety and immunogenicity of XBB.1.5-containing mRNA vaccines. Preprint at medRxiv https://doi.org/10.1101/2023.08.22.23293434 (2023).
Zhang, J. et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372, 525–530 (2021).
Parzych, E. M. et al. DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2. Nat. Commun. 13, 5886 (2022).
Zhou, T. et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 376, eabn8897 (2022).
Simon, V. et al. PARIS and SPARTA: finding the Achilles’ heel of SARS-CoV-2. mSphere 7, e0017922 (2022).
Iketani, S. et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604, 553–556 (2022).
Wang, Q. et al. Deep immunological imprinting due to the ancestral spike in the current bivalent COVID-19 vaccine. CellRep. Med. (in the press).
Liu, L., Huang, Y. & Wang, H. H. Fast and efficient template-mediated synthesis of genetic variants. Nat. Methods 20, 841–848 (2023).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Smith, D. J. et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
This study was supported by funding from the National Institutes of Health (NIH) Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) Assessment of Viral Evolution Program and through the NIH Collaborative Influenza Vaccine Innovation Center (Grant 75N93021C00014 to D.D.H.) and the NIH, National Institute of Allergy and Infectious Diseases (Contract 75N93019C00051 to A.G.). We acknowledge funding support from the National Science Foundation (Grant MCB-2032259 to H.H.W.). We thank all who contributed their data to the Global Initiative on Sharing Avian Flu Data database. We express our gratitude to D. Manthei, C. Gherasim, V. Blanc, P. Bennett-Baker, S. Sneeringer, L. Warsinske, T. Kowalski-Dobson, A. Meyers, Z. Chu, H. Kuiken, L. Barnes, A. Eckard, K. Lindsey, D. Davis, A. Rico, G. Simjanovski, M. Patel and N. Vydiswaran of the Immunity Associated with SARS-CoV-2 Study team for supplying serum samples. We acknowledge M. T. Yin and M. E. Sobieszczyk at Columbia University Medical Center for providing serum samples.
Author information
Authors and Affiliations
Contributions
Lihong Liu and D.D.H. conceived and supervised this project. Q.W. managed the project. Liyuan Liu, L.T.S., Yiming Huang, Y.Q. and H.H.W. constructed the spike expression plasmids. Q.W., J.H., R.M.Z. and Lihong Liu conducted pseudovirus neutralization assays. M.S.N. and Yaoxing Huang conducted authentic virus neutralization assays. Q.W. and Lihong Liu purified severe acute respiratory syndrome coronavirus 2 soluble spike proteins and hACE2 protein. Y.G. conducted bioinformatic analyses. Q.W., Lihong Liu, J.H., S.I. and J.Y. purified antibodies. Z.L. performed surface plasmon resonance assay. R.V., A.S.L. and A.G. provided clinical samples. Q.W., Y.G., Lihong Liu and D.D.H. analysed the results and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Lihong Liu, S.I., J.Y. and D.D.H. are inventors on a provisional patent application on 10-40 described in this manuscript, titled “Isolation, characterization, and sequences of potent and broadly neutralizing monoclonal antibodies against SARS-CoV-2 and its variants as well as related coronaviruses” (63/271,627). D.D.H. is a co-founder of TaiMed Biologics and RenBio; consultant to WuXi Biologics and Brii Biosciences; and a board director for Vicarious Surgical. A.G. serves on a scientific advisory board for Janssen Pharmaceuticals. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Spike sequence alignment of WA1 and BA.2 with BA.2.86 from human cases deposited to GISAID as of September 5, 2023.
The sequence numbering is based on WA1. Red boxes indicate the alignments of amino acids at position 16 and 670. “X”, low-quality sequencing data.
Extended Data Fig. 2 Serum neutralization of authentic BA.2.86 compared with EG.5.1.
Neutralizing ID50 titre of serum samples from “XBB breakthrough” cohort against authentic BA.2.86 and EG.5.1. The geometric mean ID50 titre (GMT) are presented above symbols. The neutralization assay limit of detection (dotted line) is 100. Statistical analysis was performed by employing Wilcoxon matched-pairs signed-rank test. GMT of BA.2.86 is around 1.2-fold (1.2X) higher than that of EG.5.1. n, sample size. dpi, days post infection.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Q., Guo, Y., Liu, L. et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature 624, 639–644 (2023). https://doi.org/10.1038/s41586-023-06750-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06750-w
This article is cited by
-
An mRNA vaccine encoding the SARS-CoV-2 receptor-binding domain protects mice from various Omicron variants
npj Vaccines (2024)
-
EG.5 (Eris) and BA.2.86 (Pirola) two new subvariants of SARS-CoV-2: a new face of old COVID-19
Infection (2024)
-
Anticipating the future of the COVID-19 pandemic: insights into the emergence of SARS-CoV-2 variant JN.1 and its projected impact on older adults
GeroScience (2024)
-
Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion
Nature Communications (2024)
-
SARS-CoV-2 Omicron subvariant BA.2.86: limited potential for global spread
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.