Abstract
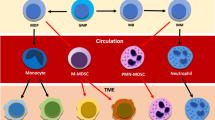
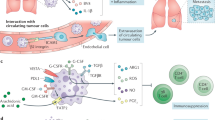
The immune-suppressive tumour microenvironment represents a major obstacle to effective immunotherapy1,2. Pathologically activated neutrophils, also known as polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), are a critical component of the tumour microenvironment and have crucial roles in tumour progression and therapy resistance2,3,4. Identification of the key molecules on PMN-MDSCs is required to selectively target these cells for tumour treatment. Here, we performed an in vivo CRISPR–Cas9 screen in a tumour mouse model and identified CD300ld as a top candidate of tumour-favouring receptors. CD300ld is specifically expressed in normal neutrophils and is upregulated in PMN-MDSCs upon tumour-bearing. CD300ld knockout inhibits the development of multiple tumour types in a PMN-MDSC–dependent manner. CD300ld is required for the recruitment of PMN-MDSCs into tumours and their function to suppress T cell activation. CD300ld acts via the STAT3-S100A8/A9 axis, and knockout of Cd300ld reverses the tumour immune-suppressive microenvironment. CD300ld is upregulated in human cancers and shows an unfavourable correlation with patient survival. Blocking CD300ld activity inhibits tumour development and has synergistic effects with anti-PD1. Our study identifies CD300ld as a critical immune suppressor present on PMN-MDSCs, being required for tumour immune resistance and providing a potential target for cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sequencing data generated in this study have been deposited to the Gene Expression Omnibus (GEO) with accession numbers GSE199601, GSE199602, GSE199603 and GSE199604. Published data used in this study were retrieved from GSE13754010. Gene associations with immune cell infiltration were obtained from TIMER 2.0 (http://timer.comp-genomics.org/timer/). The CD300ld expression profile in immune subsets was obtained from the Human Protein Atlas (https://www.proteinatlas.org/). All pre-processed TCGA metrics can be freely accessed at the Cancer Genome Atlas (TCGA) of National Cancer Institute (https://gdc.cancer.gov/). All data supporting the findings of this study are available within the Article and its Supplementary Information. Source data are provided with this paper.
References
Engblom, C., Pfirschke, C. & Pittet, M. J. The role of myeloid cells in cancer therapies. Nat. Rev. Cancer 16, 447–462 (2016).
Veglia, F., Sanseviero, E. & Gabrilovich, D. I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 21, 485–498 (2021).
Quail, D. F. et al. Neutrophil phenotypes and functions in cancer: a consensus statement. J. Exp. Med. 219, e20220011 (2022).
Jaillon, S. et al. Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer 20, 485–503 (2020).
Morad, G., Helmink, B. A., Sharma, P. & Wargo, J. A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021).
Hegde, S., Leader, A. M. & Merad, M. MDSC: markers, development, states, and unaddressed complexity. Immunity 54, 875–884 (2021).
Mantovani, A., Cassatella, M. A., Costantini, C. & Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11, 519–531 (2011).
Bronte, V. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018).
Xie, X. et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 21, 1119–1133 (2020).
Veglia, F. et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 569, 73–78 (2019).
Veglia, F. et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J. Exp. Med. 218, e20201803 (2021).
Zhou, J., Nefedova, Y., Lei, A. & Gabrilovich, D. Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin. Immunol. 35, 19–28 (2018).
Wang, P. F. et al. Prognostic role of pretreatment circulating MDSCs in patients with solid malignancies: a meta-analysis of 40 studies. Oncoimmunology 7, e1494113 (2018).
Ahn, G. O. et al. Inhibition of Mac-1 (CD11b/CD18) enhances tumor response to radiation by reducing myeloid cell recruitment. Proc. Natl Acad. Sci. USA 107, 8363–8368 (2010).
Molgora, M. et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell 182, 886–900.e17 (2020).
Ho, C. et al. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology 55, 833–845 (2012).
Passegue, E., Wagner, E. F. & Weissman, I. L. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443 (2004).
Ouzounova, M. et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat. Commun. 8, 14979 (2017).
Gungabeesoon, J. et al. A neutrophil response linked to tumor control in immunotherapy. Cell 186, 1448–1464.e20 (2023).
Masucci, M. T., Minopoli, M., Del Vecchio, S. & Carriero, M. V. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front. Immunol. 11, 1749 (2020).
Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018).
Donato, R. et al. Functions of S100 proteins. Curr. Mol. Med. 13, 24–57 (2013).
Pruenster, M., Vogl, T., Roth, J. & Sperandio, M. S100A8/A9: from basic science to clinical application. Pharmacol. Ther. 167, 120–131 (2016).
Bitsch, R. et al. STAT3 inhibitor napabucasin abrogates MDSC immunosuppressive capacity and prolongs survival of melanoma-bearing mice. J. Immunother. Cancer 10, e004384 (2022).
Su, Y. L., Banerjee, S., White, S. V. & Kortylewski, M. STAT3 in tumor-associated myeloid cells: multitasking to disrupt immunity. Int. J. Mol. Sci. 19, 1803 (2018).
Cheng, P. et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205, 2235–2249 (2008).
Hsu, K., Chung, Y. M., Endoh, Y. & Geczy, C. L. TLR9 ligands induce S100A8 in macrophages via a STAT3-dependent pathway which requires IL-10 and PGE2. PLoS ONE 9, e103629 (2014).
Nakano, T. et al. Activation of neutrophils by a novel triggering immunoglobulin-like receptor MAIR-IV. Mol. Immunol. 45, 289–294 (2008).
Kastenmuller, W., Kastenmuller, K., Kurts, C. & Seder, R. A. Dendritic cell-targeted vaccines—hope or hype? Nat. Rev. Immunol. 14, 705–711 (2014).
Nakazawa, Y. et al. Tumor-derived extracellular vesicles regulate tumor-infiltrating regulatory T cells via the inhibitory immunoreceptor CD300a. eLife 10, e61999 (2021).
Highfill, S. L. et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl Med. 6, 237ra267 (2014).
Groth, C. et al. Blocking migration of polymorphonuclear myeloid-derived suppressor cells inhibits mouse melanoma progression. Cancers 13, 726 (2021).
Condamine, T. et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J. Clin. Invest. 124, 2626–2639 (2014).
Vogl, T. et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood 104, 4260–4268 (2004).
Pruenster, M. et al. Extracellular MRP8/14 is a regulator of β2 integrin-dependent neutrophil slow rolling and adhesion. Nat. Commun. 6, 6915 (2015).
Zhao, F. et al. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology 136, 176–183 (2012).
Kinoshita, R. et al. Newly developed anti-S100A8/A9 monoclonal antibody efficiently prevents lung tropic cancer metastasis. Int. J. Cancer 145, 569–575 (2019).
Passey, R. J. et al. A null mutation in the inflammation-associated S100 protein S100A8 causes early resorption of the mouse embryo. J. Immunol. 163, 2209–2216 (1999).
Borrego, F. The CD300 molecules: an emerging family of regulators of the immune system. Blood 121, 1951–1960 (2013).
Vitalle, J. et al. The expression and function of CD300 molecules in the main players of allergic responses: mast cells, basophils and eosinophils. Int. J. Mol. Sci. 21, 3173 (2020).
Shi, L. et al. DIgR2, dendritic cell-derived immunoglobulin receptor 2, is one representative of a family of IgSF inhibitory receptors and mediates negative regulation of dendritic cell-initiated antigen-specific T-cell responses. Blood 108, 2678–2686 (2006).
Simhadri, V. R. et al. Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood 119, 2799–2809 (2012).
Choi, S. C. et al. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J. Immunol. 187, 3483–3487 (2011).
Murakami, Y. et al. CD300b regulates the phagocytosis of apoptotic cells via phosphatidylserine recognition. Cell Death Differ. 21, 1746–1757 (2014).
Haga, K. et al. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc. Natl Acad. Sci. USA 113, E6248–E6255 (2016).
Qi, S. et al. WWC proteins mediate LATS1/2 activation by Hippo kinases and imply a tumor suppression strategy. Mol. Cell 82, 1850–1864.e7 (2022).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019).
Wang, B. et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc. 14, 756–780 (2019).
Luo, M. et al. A STING-activating nanovaccine for cancer immunotherapy. Nat. Nanotechnol. 12, 648–654 (2017).
Li, C. et al. ADAP and SKAP55 deficiency suppresses PD-1 expression in CD8+ cytotoxic T lymphocytes for enhanced anti-tumor immunotherapy. EMBO Mol. Med. 7, 754–769 (2015).
Mastio, J. et al. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J. Exp. Med. 216, 2150–2169 (2019).
Acknowledgements
OT-1 TCR transgenic mice were provided by H. Wang; Mx1cre mice were provided by B. Zhou; Lyz2cre mice were provided by Z. Zhou; S100a8cre mice were provided by J. Wang; HCC-related plasmids mice were provided by F. Yu. We thank the Core Facility of SIBCB for technical help, and D. Li and G. Riddihough for discussion and revision of the paper. This study was funded by the National Natural Science Foundation of China (82341025, 32270137, 32130025, 82272142, 32293232, 81873438 and 81873922) and the National Key Research and Development Program of China 2020YFA0509000. The project was supported by Shanghai Municipal Science and Technology Major Project.
Author information
Authors and Affiliations
Contributions
M. Luo, Y.Z., H.G. and Z.L. conceived the project. C.W., X.J., J.Z., J.C. and Y.G. performed CRISPR screen experiments and analysis. C.W., X.Z. and H.G. performed subcutaneous and spontaneous tumour models and analysed the tumour microenvironment with help from J.W., Y.L., X.L. and G.S. C.W. performed in vitro cell experiments with help from X.Z., J.W., Y.L., X.L., M. Lin and C.D. G.S. performed protein purification. C.W. performed staining-related experiments with help from X.L. and J.P. C.W. and X.L. performed molecular experiments with help from P.Z. M. Lin and C.D. provided clinical samples. Z.L., M. Luo, Y.Z., J.Z. and C.W. performed bioinformatics analyses and analysed the data. M. Luo, Y.Z., Z.L., H.G. and C.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M. Luo, Y.Z. and Z.L. are listed as inventors on pending patent applications related to CD300ld. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Georgina Clark, Dmitry Gabrilovich and Ilaria Malanchi for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CD300ld expression pattern in neutrophil differentiation states.
a, RT-qPCR showing the expression of CD300ld in the indicated mouse tissues (n = 3 mice). b–d, Gating strategy of conventional DC1 (cDC1) and DC2 (cDC2), and plasmacytoid DC (pDC) in SP (b). Representative histograms (c) and statistics (d) showing the expression of CD300ld on the indicated DCs (n = 3 mice). e–g, Gating strategy of different neutrophil states in BM, SP and PB (e). Representative histograms (f) and statistics (g) showing the expression of CD300ld in the indicated neutrophil subpopulations (n = 3 mice). h: Violin plots showing the expression of CD300ld in each neutrophil subset in BM, SP and PB from naïve mice. scRNA-seq data is retrieved from Xie et al.10 (GSE137540). Data are presented as mean ± s.e.m. and are representative of at least three independent experiments (b–g).
Extended Data Fig. 2 Upregulation of CD300ld in neutrophils upon tumor-bearing.
a, Representative flow cytometry histograms (left) and statistics (right) showing the expression of CD300ld in immune cell populations from the spleen of B16-F10 tumor-bearing mice (n = 2). b, Volcano plot showing the expression comparison of membrane encoding genes in PMN-MDSCs from the spleen of B16 tumor-bearing mice versus neutrophil from the spleen of healthy mice. c, Pathway enrichment analysis with the genes significantly altered in PMN-MDSCs compared to neutrophil. d, Representative flow cytometry histograms (left) and statistics (right) showing the expression of CD300ld in monocyte and macrophage from the spleen of tumor-free mice or B16-F10 tumor-bearing mice. (n = 4). e, Representative flow cytometry histograms (left) and statistics (right) showing the expression of CD300ld in neutrophil subsets from the spleen of tumor-free or tumor-bearing mice (n = 3). f, Representative flow cytometry histograms (left) and statistics (right) showing the percent of CD14+ cells in neutrophils from tumor-free spleen or B16-F10 tumor-bearing spleen/tumor (n = 4). g, Representative flow cytometry histograms (left) and statistics (right) showing the expression of CD300ld in CD14−, CD14mid and CD14high neutrophils from B16-F10 tumors (n = 5). h, RT-qPCR analysis showing the expression of CD300ld in CD14−, CD14mid and CD14high neutrophils from B16-F10 tumors (n = 2). i, j, Representative flow cytometry histograms (i, left) and statistics (i, right, n = 4), and RT-qPCR (j, n = 3) showing the expression of CD300ld in tumor-free splenic neutrophils that were cultured in the presence of 5% B16-F10 TES or control medium for 18hrs. Each dot represents one mouse. Data are presented as mean ± s.e.m. and are representative of two (a) or at least three (d–j) independent experiments. Statistical analysis was performed using Student’s two-sided unpaired t-test (d–g, i, j).
Extended Data Fig. 3 KO of CD300ld does not alter the development of haematopoietic and immune system.
a, CRISPR-Cas9 strategy of CD300ld KO in mice. b, PCR Genotyping of CD300ld WT and KO mice. c, Representative flow cytometry plots (left) and statistics (right) showing the expression of CD300ld in neutrophils from peripheral blood of CD300ld WT and KO mice (n = 3). d,e, Representative flow cytometry plots (d) and statistics (e) showing the percentages of haematopoietic stem-cells (HSC)/progenitors in bone marrow of CD300ld WT and KO mice (n = 3). f, The colony forming ability of HSC/progenitors in bone marrow of CD300ld WT and KO mice (n = 3). g, Statistics showing the percentages of immature and mature neutrophils in bone marrow, spleen and peripheral blood of CD300ld WT and KO mice (n = 9). h,i, Flow cytometric analysis of the percentage of immune cell populations in PB of CD300ld WT and KO mice at indicated ages (n=3). Data are presented as mean ± s.e.m. Each dot represents one mouse. Data include all analyzed mice (g–i), or are representative of two (d–f) or at least three (b,c) independent experiments. Statistical analysis was performed using Student’s two-sided unpaired t-test (c, e-i).
Extended Data Fig. 4 CD300ld deletion inhibits tumor growth in a PMN-MDSCs-dependent manner.
a–c, Representative photographs (a, Bar = 1 cm) and statistics (b, WT n = 6, KO n = 5) of the tumors from WT and KO mice at day-16 post B16-F10 tumor inoculation. Panel c shows the tumor growth in individual mice. d, Tumor growth in individual mice and the mice survival after inoculated s.c with the cells of TC-1 (WT n = 6, KO n = 5), LLC (n = 7), MC38 (n = 5), or EL4 (WT n = 7, KO n = 9). e, Representative photographs of the livers from WT and KO mice at day-31 post Akt/Ras hydrodynamic injection (Bar = 1 cm, n = 5). f, TC-1 tumor growth curve in CD300ld WT and KO recipient mice that reconstituted with bone marrow cells from WT donor mice (n = 6). g,h, Strategy to generate CD300ldfl/fl mice (g) and PCR Genotyping (h). i, PCR analysis showing the conditional deletion efficiency of CD300ld in lymphocytes (T and B), Neutrophils, Monocytes and Macrophages in the indicated mice. j,k, WT mice were treated with anti-Ly6G or isotype at day-4 post B16-F10 inoculation every other day, and neutrophil presence was determined by Ly-6G intracellular staining. Neutrophil in PB at the indicated time (j, n = 5). Representative flow cytometry plots (k, left) and statistics (k, right, n = 4) showing neutrophil presence in tumor. Data are presented as mean ± s.e.m. Each dot represents one mouse. Data are representative of two (i–k) or at least three (a–h) independent experiments. Statistical analysis was performed using Student’s two-sided unpaired t-test (b,f,j,k) or log-rank test (d).
Extended Data Fig. 5 CD300ld KO remodels the tumor immune suppressive microenvironment.
a, Gating strategy to analyze immune cell populations in TME. b–e, Quantification of immune cells in tumor from the mice of Fig. 3a (b, WT n = 9, KO n = 10). The percentage of indicated immune cells in the tumor of LLC (c, n = 5), TC1 (d, n = 5) and HCC (e, n = 5) from WT and CD300ld KO mice as determined by flow cytometry. f, Gene expression from scRNA-seq experiment characterizing expression of lineage-defining genes in cell clusters. g, Key differentially expressed transcripts that distinguish cell clusters. h, Violin plots showing the expression of CD300ld in each cluster. i,j, Paired quantile–quantile plots with matched t-SNEs of CD300ld WT and KO TME, depicting the distribution of the expression of key genes involved in immune suppression of myeloid cells (i), T-cell activation/effector genes and T-cell recruitment (j). Data in b–e are presented as mean ± s.e.m. Each dot represents one mouse. Data are representative of at least three independent experiments (c–e). Statistical analysis was performed using Student’s one-sided unpaired t-test (b–e), or two-sided Wilcoxon rank-sum test (i,j).
Extended Data Fig. 6 CD300ld regulates PMN-MDSC activities.
a, GSEA profiles showing a significant enrichment of gene sets associated with neutrophil migration in WT PMN-MDSCs compared to CD300ld KO PMN-MDSCs. b, Expression levels of Cxcr2 in CD300ld WT and KO PMN-MDSCs from RNA-seq result (n = 2). c, Representative histograms (left) and statistics (right) showing the expression of CXCR2 on splenic PMN-MDSCs from CD300ld WT or KO tumor-bearing mice at day-17 post B16-F10 inoculation (n = 4). d,e, Representative plots (d) and statistics (e, n = 4) showing the gating of CTV+ PMN-MDSCs and their migration frequencies in bone marrow and tumor. f, Splenic PMN-MDSCs from B16-F10 tumor-bearing mice were isolated and cultured for 18rhs, and cell supernatants were collected for cytokine and chemokine profiling (n = 2). g, OT-I T cells were stimulated with OVA peptide in the presence of WT or CD300ld KO PMN-MDSCs. Representative plots of intracellular staining (left) and statistics (right, n = 2) are presented to show IFN-γ expression in CD8+ T cells. h–k, CD300ld WT and KO B16-F10 tumor mice were treated with anti-CD8 to deplete CD8+ T-cells. Tumor growth curve (h, WT n = 6, KO n = 5) and PMN-MDSCs recruitment in TME at day-15 post tumor inoculation (i, WT n = 6, KO n = 5) were shown. Chemotaxis of splenic PMN-MDSCs (j) and T-cell suppression of tumor derived PMN-MDSCs (k) were determined (n = 3). Data in c and h–k are presented as mean ± s.e.m. Each dot represents one mouse. Data are representative of two (f,h–k) or at least three (c,e,g) independent experiments. Statistical analysis was performed Student’s two-sided paired (e) or unpaired (c,h–k) t-test.
Extended Data Fig. 7 CD300ld is required for PMN-MDSCs migration and functions.
a, CD14+ PMN-MDSCs recruitment in B16-F10 tumor were determined by flow cytometry (WT n = 12, KO n = 9). b–d, CD300ld WT and KO mice were analyzed at day-16 post B16-F10 tumor inoculation. CD14+ cell% in PMNs (b, n = 5), Chemotaxis (c, n = 3) and T-cell suppression activity (d, n = 3) of CD14+ PMNs and CD14− PMNs derived from the tumor are presented. e-i, CD300ld WT and KO mice were analyzed at day-30 post Akt/Ras hydro-injection. CD14+ PMNs recruitment in HCC tumor was determined by flow cytometry (e, n = 5). Representative histograms (f, left) and statistics (f, right) showing the expression of CD300ld and CD14 in PMNs from HCC tumor (n = 5). CD14+ cell% in PMNs (g, n = 5), chemotaxis (h, n = 2) and T-cell suppression activity (i, n = 3) of CD14+ and CD14− PMNs derived from HCC tumor were determined. j, RNA-seq analysis showing the expression of NETosis related genes in CD300ld WT and KO PMN-MDSCs as well as the adjusted pValue (Padj) for each gene (n = 2). k, Representative Immuno-fluorescence results (k, left, Scale bars = 100 μm) showing the staining of sytox Green in CD300ld WT or KO neutrophils when treated with PMA, and the fluorescence intensities as measured by a microplate reader (k, right, n = 4). l, GSEA profiles showing a significant enrichment of neutrophils associated gene sets in CD300ld KO PMN-MDSCs compared to the WT. Data in a-k are presented as mean ± s.e.m. Each dot represents one mouse (a–j) or one replicate well (k). Data are representative of two (k) or at least three independent experiments (a–i). Statistical analysis was performed using Student’s two-sided unpaired t-test (a–g,i,k).
Extended Data Fig. 8 STAT3 plays essential role in CD300ld-S100A8/A9 axis.
a, S100A8 and S100A9 mRNA levels relative to GAPDH in PMN-MDSCs was measured using RT-qPCR (n = 3). b, Splenic PMN-MDSCs were cultured with or without 5% TES of B16-F10 tumors for 18hrs. S100A8 and S100A9 mRNA levels relative to control treatment was determined by RT-qPCR (n = 3). c, Representative flow cytometry plots showing the p-STAT3 levels in tumor-derived PMN-MDSCs. d, Splenic PMN-MDSCs were isolated from tumor-bearing mice at day-16 post B16-F10 inoculation for ChIP-qPCR to determine the recruitment of STAT3 at S100A8 and S100A9 promoter regions, non-specific primers was set as negative control (n = 3). e, Naïve splenic neutrophils were treated with coated TX69 antibody (20 ug/ml final concentration) or isotype for 18hrs. Representative flow cytometry plots for WT cells (left) and statistics for both CD300ld WT and KO cells (right) were presented to show the levels of p-STAT3 (n = 2). f, Naïve WT splenic neutrophils were treated with coated TX69 antibody (20 ug/ml final concentration) with or without STAT3 inhibitor, WP1066, at a final concentration of 10uM for 18hrs. Expression of S100A8 and S100A9 relative to isotype alone was determined using RT-qPCR (n = 3). g,h: Mice were treated with STAT3 inhibitor (1 mg/kg/day) or PBS at day-7 post B16-F10 tumor inoculation. Tumor growth curves (g, WT+PBS n = 7, WT+Stat3i n = 8, KO+PBS n = 6, KO+Stat3i n = 8) and PMN-MDSCs recruitment in tumor at day-19 post tumor inoculation (h, WT+PBS n = 6, WT+Stat3i n = 7, KO+PBS n = 6, KO+Stat3i n = 6) were shown. Data are presented as mean ± s.e.m. Each dot represents one mouse (a,e,g,h) or a replicate well (b,d,e). Data are representative of at least three independent experiments (a–f). Statistical analysis was performed using ordinary one-way ANOVA (f–h) or Student’s two-sided unpaired t-test (a,b,d).
Extended Data Fig. 9 CD300ld blockade inhibits tumor growth.
a, CD300ld expression on neutrophils from spleen of CD300ldfl/fl and CD300ldfl/flMx1cre mice that were treated with (right) or without (left) poly(I:C) (n = 4). b and c, CD300ldfl/fl and CD300ldfl/flMx1cre mice were treated with poly(I:C) for 24hrs, and DC activation in lymph node (b) or spleen (c) were determined (n = 3). d, CD300ldfl/fl and CD300ldfl/fl Mx1cre mice were treated with poly(I:C) from day-10 post B16-F10 inoculation. Tumor growth curves were shown (n = 7). e, B16-F10 cells were treated with hFc or ld-ECD proteins for 48hrs and cell viability was determined by CCK8 assay (n = 3). f, The percentage of immune subpopulations in B16-F10 tumor from the mice of Fig. 5e (n = 6). g,h, HCC mice were treated with hFc-ld-ECD or hFc proteins as described in Fig. 5g, and were analyzed at day-27 post Akt/Ras hydrodynamic injection. The photographs of liver (g, left, Bar = 1 cm), the statistics of the liver/body ratio (g, right) and the percentage of immune subpopulations in tumor (h) were shown (n = 5). i,j, hFc or hFc-ld-ECD protein was administrated into MMTV-PyMT mice. Tumor growth curve (i, left, n = 6) and mice survival (i, right, n = 6) were presented, and the percentage of immune subpopulations in breast tumor was determined at week-6 post the first treatment (j, n = 6). k, PD-1+ cell% in CD8+ T cells, and PD-L1+ cell% in PMN-MDSCs and tumor cells from B16-F10 tumor (WT n = 6, KO n = 5). Data are presented as mean ± s.e.m. Each dot represents one mouse. Data are representative of two (b,c,e,i–j) or at least three independent experiments (a,d–f). Statistical analysis was performed using Student’s two-sided unpaired t-test (a–d,f–h,i-left, j,k) or log-rank test (i-right).
Extended Data Fig. 10 Conserved function of CD300LD in human cancers.
a, Expression of CD300LD in human immune cells with the data from Human Protein Atlas. b, Representative Immuno-fluorescence results showing the CD300LD antibody staining of 293t cells transfected with individual CD300 family members, Scale bars = 10 μm. c, Representative Immuno-fluorescence results showing the staining of MPO, CD15 and CD300LD in human melanoma samples. Scale bars = 10 μm. d, Correlation analysis of CD300LD expression with PMN-MDSCs infiltration in human cancers by TIMER 2.0. e, CRISPR-Cas9 strategy to generate CD300ld humanized (HuLD) mice. f, PCR Genotyping of HuLD mice. g, mRNA levels of human CD300LD relative to GAPDH in blood cells from WT, CD300ld KO and huLD mice were measured by RT-qPCR (n = 3). h, Survival curve of mice after inoculated s.c with the cells of B16-F10 (WT n = 7, HuLD n = 7, KO n = 6). i, TC-1 tumor growth curves in WT and huLD mice (WT n = 6, HuLD n = 4). j, Survival curve of mice after inoculated s.c with B16-F10 and then treated with hFc or LD-ECD protein (n = 10). k, CD300ld acts as an immune suppressor on PMN-MDSCs for tumor-driven immune suppression. Data are presented as mean ± s.e.m. Data are representative of two (b,c,f and g) or at least three independent experiments (h–j). Statistical analysis was performed using Student’s two-sided unpaired t-test (g,i) or log-rank test (h,j).
Supplementary information
Supplementary Table 1
Screen gene summary. Gene summary of in vivo CRISPR–Cas9 screening (bone marrow versus tumour).
Supplementary Table 2
Screen sgRNA summary. sgRNA summary of in vivo CRISPR–Cas9 screening (bone marrow versus tumour).
Supplementary Table 3
RNA-seq neutrophil vs PMN-MDSC. RNA-seq comparison between neutrophils from tumour-free and tumour-bearing mice.
Supplementary Table 4
RNA-seq PMN-MDSC KO vs wild type. RNA-seq comparison between wild-type and Cd300ld-KO PMN-MDSCs.
Supplementary Table 5
Primers used for RT–qPCR analyses.
Supplementary Table 6
Signature genes. CD300ld-related signatures genes used in cancer patient survival analyses.
Supplementary Table 7
Summary information of cancer patients and healthy volunteers.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, C., Zheng, X., Zhang, J. et al. CD300ld on neutrophils is required for tumour-driven immune suppression. Nature 621, 830–839 (2023). https://doi.org/10.1038/s41586-023-06511-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06511-9
This article is cited by
-
CRISPR–Cas9 applications in T cells and adoptive T cell therapies
Cellular & Molecular Biology Letters (2024)
-
“Moderate” adjuvant chemotherapy-induced leukopenia is beneficial for survival of patients with early breast cancer: a retrospective study
BMC Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.