Abstract
Skeletal muscle atrophy is a hallmark of the cachexia syndrome that is associated with poor survival and reduced quality of life in patients with cancer1. Muscle atrophy involves excessive protein catabolism and loss of muscle mass and strength2. An effective therapy against muscle wasting is currently lacking because mechanisms driving the atrophy process remain incompletely understood. Our gene expression analysis in muscle tissues indicated upregulation of ectodysplasin A2 receptor (EDA2R) in tumour-bearing mice and patients with cachectic cancer. Here we show that activation of EDA2R signalling promotes skeletal muscle atrophy. Stimulation of primary myotubes with the EDA2R ligand EDA-A2 triggered pronounced cellular atrophy by induction of the expression of muscle atrophy-related genes Atrogin1 and MuRF1. EDA-A2-driven myotube atrophy involved activation of the non-canonical NFĸB pathway and was dependent on NFκB-inducing kinase (NIK) activity. Whereas EDA-A2 overexpression promoted muscle wasting in mice, deletion of either EDA2R or muscle NIK protected tumour-bearing mice from loss of muscle mass and function. Tumour-induced oncostatin M (OSM) upregulated muscle EDA2R expression, and muscle-specific oncostatin M receptor (OSMR)-knockout mice were resistant to tumour-induced muscle wasting. Our results demonstrate that EDA2R–NIK signalling mediates cancer-associated muscle atrophy in an OSM–OSMR-dependent manner. Thus, therapeutic targeting of these pathways may be beneficial in prevention of muscle loss.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Human gene expression datasets analysed in this study are available in the GEO database: GSE130563 (ref. 34), GSE34111 (ref. 35), GSE1007 (ref. 36), GSE115650 (ref. 37) and GSE140261 (ref. 38). A detailed description of muscle biopsies from patients with cancer used in this study was previously published31. Source Data are provided with this paper.
References
Baracos, V. E., Martin, L., Korc, M., Guttridge, D. C. & Fearon, K. C. H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 4, 17105 (2018).
Cohen, S., Nathan, J. A. & Goldberg, A. L. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 14, 58–74 (2015).
Argiles, J. M., Stemmler, B., Lopez-Soriano, F. J. & Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol. 15, 9–20 (2018).
Dolly, A., Dumas, J. F. & Servais, S. Cancer cachexia and skeletal muscle atrophy in clinical studies: what do we really know? J. Cachexia Sarcopenia Muscle 11, 1413–1428 (2020).
Kowalczyk-Quintas, C. & Schneider, P. Ectodysplasin A (EDA) – EDA receptor signalling and its pharmacological modulation. Cytokine Growth Factor Rev. 25, 195–203 (2014).
Yan, M. et al. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science 290, 523–527 (2000).
Sadier, A., Viriot, L., Pantalacci, S. & Laudet, V. The ectodysplasin pathway: from diseases to adaptations. Trends Genet. 30, 24–31 (2014).
Newton, K., French, D. M., Yan, M., Frantz, G. D. & Dixit, V. M. Myodegeneration in EDA-A2 transgenic mice is prevented by XEDAR deficiency. Mol. Cell. Biol. 24, 1608–1613 (2004).
Das, S. K. et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 333, 233–238 (2011).
Kir, S. et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature 513, 100–104 (2014).
Sinha, S. K., Zachariah, S., Quinones, H. I., Shindo, M. & Chaudhary, P. M. Role of TRAF3 and -6 in the activation of the NF-kappa B and JNK pathways by X-linked ectodermal dysplasia receptor. J. Biol. Chem. 277, 44953–44961 (2002).
Verhelst, K. et al. XEDAR activates the non-canonical NF-kappaB pathway. Biochem. Biophys. Res. Commun. 465, 275–280 (2015).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. https://doi.org/10.1038/sigtrans.2017.23 (2017).
Awazawa, M. et al. A microRNA screen reveals that elevated hepatic ectodysplasin A expression contributes to obesity-induced insulin resistance in skeletal muscle. Nat. Med. 23, 1466–1473 (2017).
Valino-Rivas, L. et al. NIK as a druggable mediator of tissue injury. Trends Mol. Med. 25, 341–360 (2019).
Dogra, C. et al. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 21, 1857–1869 (2007).
Johnston, A. J. et al. Targeting of Fn14 prevents cancer-induced cachexia and prolongs survival. Cell 162, 1365–1378 (2015).
Ren, X. et al. A small-molecule inhibitor of NF-kappaB-inducing kinase (NIK) protects liver from toxin-induced inflammation, oxidative stress, and injury. FASEB J. 31, 711–718 (2017).
Li, X. et al. Activation of NF-kappaB-inducing kinase in islet beta cells causes beta cell failure and diabetes. Mol. Ther. 28, 2430–2441 (2020).
Richards, C. D. The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm. 2013, 512103 (2013).
Miki, Y. et al. Oncostatin M induces C2C12 myotube atrophy by modulating muscle differentiation and degradation. Biochem. Biophys. Res. Commun. 516, 951–956 (2019).
Webster, J. M., Kempen, L., Hardy, R. S. & Langen, R. C. J. Inflammation and skeletal muscle wasting during cachexia. Front. Physiol. 11, 597675 (2020).
Jatoi, A. et al. A placebo-controlled double blind trial of etanercept for the cancer anorexia/weight loss syndrome: results from N00C1 from the North Central Cancer Treatment Group. Cancer 110, 1396–1403 (2007).
Jatoi, A. et al. A placebo-controlled, double-blind trial of infliximab for cancer-associated weight loss in elderly and/or poor performance non-small cell lung cancer patients (N01C9). Lung Cancer 68, 234–239 (2010).
Bayliss, T. J., Smith, J. T., Schuster, M., Dragnev, K. H. & Rigas, J. R. A humanized anti-IL-6 antibody (ALD518) in non-small cell lung cancer. Expert Opin. Biol. Ther. 11, 1663–1668 (2011).
Marceca, G. P., Londhe, P. & Calore, F. Management of cancer cachexia: attempting to develop new pharmacological agents for new effective therapeutic options. Front. Oncol. 10, 298 (2020).
Honors, M. A. & Kinzig, K. P. The role of insulin resistance in the development of muscle wasting during cancer cachexia. J. Cachexia Sarcopenia Muscle 3, 5–11 (2012).
Barbera, M. C. et al. Increased expression of ectodysplasin A2 receptor EDA2R is the most remarkable and ubiquitous aging-related transcriptional hallmark. Preprint at https://doi.org/10.21203/rs.3.rs-774469/v1 (2021).
Brightbill, H. D. et al. Conditional deletion of NF-kappaB-inducing kinase (NIK) in adult mice disrupts mature B cell survival and activation. J. Immunol. 195, 953–964 (2015).
Springer, M. L., Rando, T. A. & Blau, H. M. Gene delivery to muscle. Curr. Protoc. Hum. Genet. https://doi.org/10.1002/0471142905.hg1304s31 (2002).
Loumaye, A. et al. Role of activin A and myostatin in human cancer cachexia. J. Clin. Endocrinol. Metab. 100, 2030–2038 (2015).
Fearon, K. et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 12, 489–495 (2011).
Gueugneau, M. et al. Increased Serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. J. Cachexia Sarcopenia Muscle 9, 929–946 (2018).
Judge, S. M. et al. Skeletal muscle fibrosis in pancreatic cancer patients with respect to survival. JNCI Cancer Spectr. 2, pky043 (2018).
Gallagher, I. J. et al. Suppression of skeletal muscle turnover in cancer cachexia: evidence from the transcriptome in sequential human muscle biopsies. Clin. Cancer Res. 18, 2817–2827 (2012).
Haslett, J. N. et al. Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics 4, 163–171 (2003).
Wang, L. H. et al. MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum. Mol. Genet. 28, 476–486 (2019).
Wong, C. J. et al. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum. Mol. Genet. 29, 1030–1043 (2020).
Acknowledgements
We gratefully acknowledge the use of the animal facility infrastructure at Koc University Research Centre for Translational Medicine. We thank M. Kracht (JLU Giessen) for sharing human NIK plasmids and S.-C. Sun (MD Anderson Cancer Center) for sharing NIK-floxed mice. EDA2R-null and NIK-floxed mice were provided by Genentech. We also thank I. Oguz for assistance. D.H.A. was funded by a TUBITAK-BIDEB scholarship. This work was supported by the Scientific and Technological Research Council of Turkey (grant nos. 118Z167, 118Z791, 118C014 and 122Z163) and by EMBO Installation Grant no. 4162 to S.K.
Author information
Authors and Affiliations
Contributions
S.K. conceived and designed the experiments. S.N.B., A.D. and S.K. performed myotube culture experiments, including RT-qPCR analysis and immunoblotting. S.N.B. and A.D. performed fluorescence microscopy imaging of myotubes and measurement of their diameter. With the assistance of D.H.A., A.D. performed MyHC staining of myotubes and intensity measurements. A.D. conducted immunohistochemistry staining. B.T., S.K., B.Z.C.W. and S.N.B. prepared adenovirus vectors. B.T. and S.K. purified adenoviruses and performed cell culture and mouse experiments involving adenoviruses, including RT-qPCR analysis and immunoblotting. S.N.B. and S.K. performed in vitro phosphatase assays and site-directed mutagenesis studies. S.N.B., A.D. and S.K. conducted tumour inoculation experiments, tissue collection and grip strength measurements. B.Z.C.W. contributed to tissue harvests. S.N.B. and A.D. performed RT-qPCR analysis and immunoblotting of tissue samples. S.N.B. and A.D. conducted tissue histology and S.A. performed quantification of sections. Z.O. performed genotyping of all transgenic mice. J.-P.T. and A.L. supervised and performed muscle biopsy collection from patients with cancer. P.L. performed cDNA library preparation from RNA samples of muscle biopsies. S.N.B. performed qPCR analysis of human cDNA. S.A. conducted all statistical analyses and analysis of gene expression datasets. S.N.B., A.D. and S.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Andreas Bergthaler, Pascal Schneider and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 The expression of Eda-a2 and Eda2r is enriched in skeletal muscle tissue.
a-d, Various tissue samples were collected from C57BL/6 mice. Relative mRNA levels were determined by RT-qPCR (n = 4 mice). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two independent experiments (a–d).
Extended Data Fig. 2 EDA2R expression is induced in DMD and FSHD patients.
a, EDA2R transcript levels were analyzed in quadriceps muscle biopsies collected from cachectic patients with upper gastrointestinal cancer (n = 12) before and after tumor resection (GSE34111). Upon surgery, EDA2R levels were upregulated in 4 patients (red connecting lines) and downregulated in 8 patients (green connecting lines). n.s. statistically not significant. b, GSE1007 dataset was analyzed by GEO2R and EDA2R expression values were determined in normal subjects and DMD patients (n = 10). In each group, one individual had 2 technical replicates making the total number of data points 11. c, GSE115650 dataset was analyzed by DESeq2 and EDA2R expression values were determined in normal subjects (n = 9) and FSHD patients (n = 34). d, GSE140261 dataset was analyzed by DESeq2 and EDA2R expression values were determined in normal subjects (n = 8) and FSHD patients (n = 27). The values are mean ± SEM. Statistical analysis was conducted using the two-tailed paired t-test (a). Adjusted P values were calculated with Benjamini & Hochberg false discovery rate method by GEO2R (b,c) and DESeq2 (d).
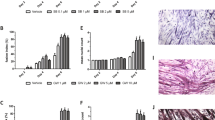
Extended Data Fig. 3 The overexpression of EDA-A2 or the administration of recombinant EDA-A2 in human and mouse myotubes stimulates cellular atrophy.
a–c, Mouse primary myotubes were transduced with LacZ, EDA-A1, or EDA-A2 expressing adenoviruses. 24 hr later, gene expression was tested by RT-qPCR (n = 3) (a). Myotubes also treated with GFP adenovirus were visualized 48 hr later under the fluorescence microscope. The scale bar is 50 µm (b). Average myotube diameter was measured (n = 3) (c). d–f, Human Skeletal Muscle Myoblasts (HSMM) were differentiated into myotubes and treated with an adenovirus expressing EDA-A2 (d) or recombinant EDA-A2 protein (250 ng/ml) (e,f) for 48 hr. Gene expression was determined by RT-qPCR (n = 3) (d,e). Human myotubes were investigated under the light microscope. The scale bar is 100 µm (f). g,h, Mouse primary myotubes were treated with recombinant EDA-A1, EDA-A2 or TNFα proteins (250 ng/ml each) for 48 hr. MyHC was immunofluorescently labeled while nuclei was counterstained with DAPI. The scale bar is 50 µm (g). MyHC signal was normalized to the number of myotube nuclei (n = 3) (h). i, Mouse primary myotubes treated with recombinant EDA-A2 (250 ng/ml) and proteasome inhibitor MG132 (10 µM) were lysed and protein samples were studied by western blotting. The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two (a–d,f–i) or three (e) independent experiments. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (a,c,h) or the two-tailed unpaired t-test (d,e).
Extended Data Fig. 4 EDA-A2 stimulates the expression of NFĸB signaling components and the alternative NFĸB activation in primary myotubes. Electrophoretic mobility shift of mouse NIK protein depends on its autophosphorylation and kinase activity.
a,b, Mouse primary myotubes were treated with recombinant EDA-A2 (250 ng/ml) for 24 hr and gene expression was determined by RT-qPCR (n = 3). c,d, Mouse primary myotubes were transduced with adenoviruses expressing LacZ, wild-type mouse NIK (mNIK), autophosphorylation-deficient mNIK-T561A mutant and kinase-dead mNIK-K431/432A mutant or human NIK (hNIK). A day later, recombinant EDA-A2 (250 ng/ml) was also added for another 24 hr. Protein levels were determined by western blotting. e, Mouse primary myotubes were transduced with LacZ, EDA-A1, or EDA-A2 expressing adenoviruses. 24 hr later, protein levels were determined by western blotting. The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two (b,c,e) or three (a,d) independent experiments. Statistical analysis was conducted using the two-tailed unpaired t-test.
Extended Data Fig. 5 Activation of the canonical NFĸB signaling is dispensable for EDA-A2-induced gene expression in primary myotubes.
a,b, Mouse primary myotubes were treated with IĸB phosphorylation inhibitors BAY 11-7082 (10 µM) and BOT-64 (10 µM) in combination with recombinant EDA-A2 (250 ng/ml) for 24 hr. Gene expression was studied by RT-qPCR (n = 3) (a) and protein levels were determined by western blotting (b). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two independent experiments. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test.
Extended Data Fig. 6 Overexpression of NIK promotes the alternative NFĸB activation and atrophy in primary myotubes.
a,b, Mouse primary myotubes were transduced with adenoviruses expressing LacZ or human NIK (hNIK). 24 hr later, protein levels were determined by western blotting (a). Myotubes also treated with GFP adenovirus were visualized 48 hr later under the fluorescence microscope and average myotube diameter was measured (n = 3) (b). c–e, Human Skeletal Muscle Myoblasts (HSMM) were differentiated into myotubes and treated with adenoviruses expressing LacZ or human hNIK for 48 hr. Gene expression was determined by RT-qPCR (n = 3) (c). Human myotubes were investigated under the light microscope. The scale bar is 100 µm (d). Myotube diameter was measured (n = 3) (e). f,g, Primary myotubes were transduced with adenoviruses expressing LacZ, mouse NIK (mNIK), autophosphorylation-deficient mNIK-T561A mutant or kinase dead mNIK-K431/432A mutant. Protein levels were determined by western blotting (f). This is the same experiment as Extended Data Fig. 4a. NIK and p65-RelA blots were cropped from Extended Data Fig. 4a. mRNA levels were tested by RT-qPCR (n = 3) (g). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two (a,b,d–g) or three (c) independent experiments. Statistical analysis was conducted using the two-tailed unpaired t-test (b,c,e) and one-way ANOVA with Tukey’s multiple-comparison test (g).
Extended Data Fig. 7 The inhibition of NIK kinase activity with B022 or a dominant-negative NIK mutant blocks EDA-A2’s effects in primary myotubes.
a,b, Mouse primary myotubes were transduced with adenoviruses expressing LacZ or human NIK and treated with different doses of B022 (1 µM, 5 µM or 10 µM) for 24 hr. Protein levels were determined by western blotting (a) and changes in gene expression were tested by RT-qPCR (n = 3) (b). c, Mouse primary myotubes were transduced with LacZ or EDA-A2 adenoviruses and treated with B022 (5 µM) for 24 hr. Protein levels were determined by western blotting. d, Mouse primary myotubes were transduced with adenoviruses expressing wild-type human NIK or the dominant-negative human NIK-K429/430A mutant. Protein levels were determined by western blotting. e,f, Mouse primary myotubes were transduced with adenoviruses expressing LacZ or the NIK-K429/430A mutant and treated with recombinant EDA-A2 (100 ng/ml) for 24 hr. Protein levels were determined by western blotting (e) and changes in gene expression were tested by RT-qPCR (n = 3) (f). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two (a–d,f) or three (e) independent experiments. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test.
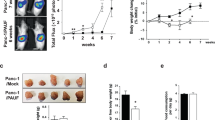
Extended Data Fig. 8 EDA2R-deficient mice are resistant to tumor-induced muscle wasting.
a-d,j, Mice were inoculated with LLC cells and sacrificed 16 days later (n = 6 mice per group). Carcass weight without the tumor mass (a) and tumor weight (b) were measured. c, Gene expression levels in tumor samples were measured by RT-qPCR (n = 6 mice). d, Plasma CRP levels were determined by ELISA (n = 6 mice). e–i, k–l, Mice were inoculated with B16 cells and sacrificed 14 days later (EDA2R-KO-B16 n = 5, other groups n = 6 mice). Carcass weight without the tumor mass (e) and tumor weight (f) were measured. A decrease in carcass weight was induced by LLC tumors. However, tissue wasting was not reflected in the carcass weight when mice received B16 tumors. Because these tumors cause excessive subcutaneous swelling due to inflammation which masks the wasting. g–i, Gastrocnemius muscle cross-sections were H&E stained (g), cross-sectional area (h) and the fiber frequency distribution (i) were measured. The scale bar is 100 µm. j, Quadriceps muscle mRNA levels of the LLC tumor-bearing mice were tested by RT-qPCR (n = 6 mice). k,l, Gastrocnemius muscle (k) and quadriceps muscle (l) mRNA levels of the B16 tumor-bearing mice were determined by RT-qPCR (EDA2R-KO-B16 n = 5, other groups n = 6 mice). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of three independent experiments (a–l). Statistical analysis was conducted using two-way ANOVA with Tukey’s multiple-comparison test (a,h,j,k,l).
Extended Data Fig. 9 Muscle-specific depletion of NIK protects from tumor-induced muscle wasting.
a. Nik mRNA levels were tested by RT-qPCR in various tissues of the Myo-NIK-KO mice (n = 3 mice). b–k, Mice were inoculated with LLC cells and sacrificed 16 days later (Myo-NIK-KO-LLC n = 5, other groups n = 6 mice). Carcass weight without the tumor mass (b) and tumor weight (c) were measured. Collected tissues were weighed (d). Forelimb grip strength was measured before the sacrifice (e). f–h, Gastrocnemius muscle cross-sections were H&E stained (f), cross-sectional area (g) and the fiber frequency distribution (h) were measured. The scale bar is 100 µm. i,j, Gastrocnemius muscle mRNA levels were tested by RT-qPCR (Myo-NIK-KO-LLC n = 5, other groups n = 6 mice) (i) and their protein levels were determined by western blotting (n = 3 mice) (j). Quadriceps muscle mRNA levels were tested by RT-qPCR (Myo-NIK-KO-LLC n = 5, other groups n = 6 mice) (k). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of three independent experiments (a–k). Statistical analysis was conducted using the two-tailed unpaired t-test (a) or two-way ANOVA with Tukey’s multiple-comparison test (b,d,e,g,i,k).
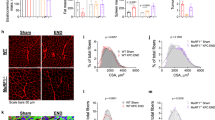
Extended Data Fig. 10 OSM induces Eda2r expression in muscle and the depletion of OSMR protects from muscle wasting.
a, Mouse primary myotubes were treated with recombinant TNFα, IL-6, LIF and OSM (250 ng/ml each). mRNA levels were determined by RT-qPCR (n = 3). b,c, Mouse primary myotubes were treated with recombinant OSM and EDA-A2 (250 ng/ml each) for 48 hr. Myotubes also treated with the GFP adenovirus were visualized under the fluorescence microscope. The scale bar is 50 µm (b). Average myotube diameter was measured (n = 3) (c). d, Mouse primary myotubes were treated with recombinant OSM (250 ng/ml for 48 hr) and EDA-A2 (100 ng/ml for 24 hr). Changes in gene expression were determined by RT-qPCR (n = 3). e–h, Mice were inoculated with LLC cells and sacrificed 16 days later (WT-LLC n = 5, other groups n = 6 mice). Carcass weight without the tumor mass (e) and tumor weight (f) were measured. Collected adipose tissues were weighed (g). Quadriceps muscle mRNA levels were tested by RT-qPCR (WT-LLC n = 5, other groups n = 6 mice) (h). The values are mean ± SEM. Each dot represents a biological replicate. Data are representative of two (a–d) or three (e–h) independent experiments. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (a,c,d) and two-way ANOVA with Tukey’s multiple-comparison test (h).
Supplementary information
Supplementary Fig. 1
Uncropped immunoblot images with molecular weight markers are shown for Figs. 1k–m, 2b–d, 3b, 4i and 5k, and for Extended Data Figs. 3i, 4c–e, 5b, 6a,f, 7a,c–e and 9j.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bilgic, S.N., Domaniku, A., Toledo, B. et al. EDA2R–NIK signalling promotes muscle atrophy linked to cancer cachexia. Nature 617, 827–834 (2023). https://doi.org/10.1038/s41586-023-06047-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06047-y
This article is cited by
-
Ectodysplasin A2 receptor-NF-κB-inducing kinase axis: a new player in muscle wasting to cancer cachexia
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.