Abstract
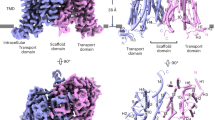
The PIN-FORMED (PIN) protein family of auxin transporters mediates polar auxin transport and has crucial roles in plant growth and development1,2. Here we present cryo-electron microscopy structures of PIN3 from Arabidopsis thaliana in the apo state and in complex with its substrate indole-3-acetic acid and the inhibitor N-1-naphthylphthalamic acid (NPA). A. thaliana PIN3 exists as a homodimer, and its transmembrane helices 1, 2 and 7 in the scaffold domain are involved in dimerization. The dimeric PIN3 forms a large, joint extracellular-facing cavity at the dimer interface while each subunit adopts an inward-facing conformation. The structural and functional analyses, along with computational studies, reveal the structural basis for the recognition of indole-3-acetic acid and NPA and elucidate the molecular mechanism of NPA inhibition on PIN-mediated auxin transport. The PIN3 structures support an elevator-like model for the transport of auxin, whereby the transport domains undergo up–down rigid-body motions and the dimerized scaffold domains remain static.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Structure coordinates and cryo-EM density maps have been deposited at the Protein Data Bank and Electron Microscopy Data Bank under accession numbers 7WKS and EMD-32568 for PIN3apo; 7WKW and EMD-32570 for PIN3NPA; and 7XXB and EMD-33500 for PIN3IAA. Other structure coordinates analysed in the paper can be downloaded from Protein Data Bank under accession numbers 4N7W for Y. frederiksenii ASBT; 3ZUY for Neisseria meningitidis ASBT; 7DSW for human NHE1; 5BZ2 for Thermus thermophilus NapA; and 4CZB for Methanocaldococcus jannaschii NhaP. Additional data supporting the findings in this study are provided in the Supplementary Information. Uncropped images for Extended Data Figs. 2c, 3a,f,k and 9d are provided in Supplementary Fig. 1. Source data are provided with this paper.
Change history
21 September 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05360-2
References
Adamowski, M. & Friml, J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32 (2015).
Krecek, P. et al. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 10, 249 (2009).
Cooke, T. J., Poli, D., Sztein, A. E. & Cohen, J. D. Evolutionary patterns in auxin action. Plant Mol. Biol. 49, 319–338 (2002).
Ljung, K., Bhalerao, R. P. & Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474 (2001).
Ljung, K. et al. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104 (2005).
Galweiler, L. et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998).
Bennett, M. J. et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950 (1996).
Noh, B., Murphy, A. S. & Spalding, E. P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13, 2441–2454 (2001).
Petrasek, J. et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 (2006).
Friml, J. et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865 (2004).
Michniewicz, M. et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056 (2007).
Huang, F. et al. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22, 1129–1142 (2010).
Zhang, J., Nodzynski, T., Pencik, A., Rolcik, J. & Friml, J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl Acad. Sci. USA 107, 918–922 (2010).
Barbosa, I. C. R., Zourelidou, M., Willige, B. C., Weller, B. & Schwechheimer, C. D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell 29, 674–685 (2014).
Zourelidou, M. et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3, e02860 (2014).
Friml, J., Wisniewska, J., Benkova, E., Mendgen, K. & Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002).
Muller, A. et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911 (1998).
Friml, J. et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673 (2002).
Mravec, J. et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140 (2009).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Teale, W. & Palme, K. Naphthylphthalamic acid and the mechanism of polar auxin transport. J. Exp. Bot. 69, 303–312 (2018).
Geisler, M., Aryal, B., di Donato, M. & Hao, P. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 58, 1601–1614 (2017).
Bailly, A. et al. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283, 21817–21826 (2008).
Titapiwatanakun, B. et al. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 57, 27–44 (2009).
Wu, G., Otegui, M. S. & Spalding, E. P. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 22, 3295–3304 (2010).
Zhu, J. et al. TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28, 930–948 (2016).
Abas, L. et al. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc. Natl Acad. Sci. USA 118, e2020857118 (2021).
Teale, W. D. et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 40, e104416 (2021).
Nodzynski, T. et al. Enquiry into the topology of plasma membrane-localized PIN auxin transport components. Mol. Plant 9, 1504–1519 (2016).
Dong, Y. et al. Structure and mechanism of the human NHE1–CHP1 complex. Nat. Commun. 12, 3474 (2021).
Winklemann, I. et al. Structure and elevator mechanism of the mammalian sodium/proton exchanger NHE9. EMBO J. 39, e105908 (2020).
Paulino, C., Wohlert, D., Kapotova, E., Yildiz, O. & Kuhlbrandt, W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. eLife 3, e03583 (2014).
Wohlert, D., Kuhlbrandt, W. & Yildiz, O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. eLife 3, e03579 (2014).
Coincon, M. et al. Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters. Nat. Struct. Mol. Biol. 23, 248–255 (2016).
Hunte, C. et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005).
Lee, C. et al. A two-domain elevator mechanism for sodium/proton antiport. Nature 501, 573–577 (2013).
Hu, N. J., Iwata, S., Cameron, A. D. & Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408–411 (2011).
Zhou, X. et al. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505, 569–573 (2014).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Ma, Q., Grones, P. & Robert, S. Auxin signaling: a big question to be addressed by small molecules. J. Exp. Bot. 69, 313–328 (2018).
Hayashi, K. I. Chemical biology in auxin research. Cold Spring Harb. Perspect. Biol. 13, a040105 (2021).
Drew, D. & Boudker, O. Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016).
Michniewicz, M., Brewer, P. B. & Friml, J. I. Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 5, e0108 (2007).
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J. & Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684 (1991).
Glanc, M. et al. AGC kinases and MAB4/MEL proteins maintain PIN polarity by limiting lateral diffusion in plant cells. Curr. Biol. 31, 1918–1930 (2021).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
The PyMOL molecular graphics system v.1.8 (Schrodinger, 2015).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Kim, S. et al. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 38, 1879–1886 (2017).
Abraham, M. J., Murtola, T., Schulz, R., Páll, S. & Lindahl, E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex 1–2, 19–25 (2015).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
Single-particle cryo-EM data were collected at the Cryo-Electron Microscopy Facility of Hubei University, Center of Cryo-Electron Microscopy at Zhejiang University, and the National Facility for Protein Science in Shanghai (NFPS). We thank X. Zhang for his support in cryo-EM facility access and data acquisition and Q. Zhang for his support in fluorescence imaging; the staff at the Radiation Technology Platform of Zhejiang University for radioactive experiments; and the staff members of the Large-scale Protein Preparation System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, Shanghai Advanced Research Institute, Chinese Academy of Science, China for providing technical support and assistance in ITC data collection and analysis. This work was supported in part by the Ministry of Science and Technology (2020YFA0908501 and 2018YFA0508100 to J.G., and 2020YFA0908400 to S.W.), the National Natural Science Foundation of China (31870724 to J.G., 31741067 and 31800990 to F. Yang, and 31900930 to S.W.), China Postdoctoral Science Foundation (2020M672434 to S.W. and 2021M692818 to N.S.), the Fundamental Research Funds for the Central Universities (2021FZZX001-28 to J.G.) and Zhejiang Provincial Natural Science Foundation (LR19C050002 to J.G. and LR20C050002 to F. Yang). J.G. and F. Yang are supported by MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University. F. Yang is supported by Alibaba Cloud.
Author information
Authors and Affiliations
Contributions
J.G., F. Yang and S.W. conceived and supervised the project. N.S., X.T., S.C., F. Ye, Y.Z., C.Z., Q.C., J.W. and C.Y.Z. performed sample preparation, data acquisition and structure determination. N.S., A.Z. and S.Z. performed the auxin efflux assay. N.S., Y.G. and S.J. performed the SPR assay. F. Ye, H.W. and F. Yang performed the molecular dynamics simulations. J.G., S.W., F. Yang, S.J.Z., S.Y., L.M. and Z.J.D. performed structure data analysis. All of the authors participated in the data analysis and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Dolf Weijers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sequence alignment of Arabidopsis thaliana PINs (AtPIN1–8).
Secondary structure assignments are based on the structure of AtPIN3apo. The coloured bars mark the locations of transmembrane helices in AtPIN3. Amino acid residues involved in IAA and NPA binding are indicated by triangles in blue. Circles indicate key residues involved in dimerization. Amino acid residues involved in CTD function are indicated by circles in orange.
Extended Data Fig. 2 IAA transport activity of AtPIN3.
a, The dose response curve of NPA inhibition on the 3H-IAA transport assay (n = 6 independent experiments). The maximum NPA inhibition is ~59%. Data were presented as mean ± s.e.m. b, The 3H-IAA efflux of WT and mutant PIN3 at 10 min in the IAA loading phase in the absence or presence of 50 µM NPA (n = 6 independent experiments). Data were presented as mean ± s.e.m. Two-tailed t-test was performed. c, The Western blot shows that all mutants are expressed at comparable levels as the WT PIN3. n = 3 independent experiments. d, Representative fluorescent (upper) and bright-field (lower) images of cells expressing green fluorescent protein (GFP)-tagged WT PIN3 and mutants. GFP fluorescence was readily detected in WT and mutant PIN3 cells with similar patterns. n = 3 independent experiments.
Extended Data Fig. 3 Structure determination of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA.
a, f, k, Size-exclusion chromatography of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA on Superose 6 (GE Healthcare) and SDS-PAGE analysis of the final sample, respectively. n = 3 independent experiments. b, g, l, Flowchart of image processing for AtPIN3apo, AtPIN3NPA, and AtPIN3IAA particles, respectively. c, h, m, The density map of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA coloured by local resolution. d, i, n, Angular distribution plot of particles included in the final C2-symmetric 3D reconstruction of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA, respectively. e, j, o, The Gold standard Fourier Shell Correlation (FSC) curve of the final 3D reconstruction of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA, and the FSC curve for cross-validation between the map and the model of AtPIN3apo, AtPIN3NPA, and AtPIN3IAA, respectively.
Extended Data Fig. 4 Sample maps of AtPIN3 structures.
a, Sample maps at 10 transmembrane helices (TMs) of AtPIN3apo. The density is shown as a mask around each TM at the level of 0.0183 in UCSF Chimera. The maps are low-pass filtered to 3.0 Å and sharpened with a B factor of −90 Å2. b, Sample maps at 10 transmembrane helices (TMs) of AtPIN3NPA. The density is shown as a mask around each TM at the level of 0.0177 in UCSF Chimera. The maps are low-pass filtered to 2.62 Å and sharpened with a B factor of −92 Å2. c, Sample maps at 10 transmembrane helices (TMs) of AtPIN3IAA. The density is shown as a mask around each TM at the level of 0.022 in UCSF Chimera. The maps are low-pass filtered to 2.93 Å and sharpened with a B factor of −70 Å2. d, Density around the substrate-binding site in AtPIN3apo (left), AtPIN3NPA (middle), and AtPIN3IAA (right) at the contour level of around 7 σ in Coot. The dashed circle marks no density in the substrate-binding site in the map of AtPIN3apo. Numbers indicate the transmembrane helices.
Extended Data Fig. 5 Structure comparisons of AtPIN3apo, AtPIN3IAA, and AtPIN3NPA.
a, Superimposition of AtPIN3apo and AtPIN3IAA subunit A structures in different orientations. The root mean square deviation (r.m.s.d.) for 377 Cα atoms in two structures is 0.90 Å. b, Superimposition of AtPIN3apo and AtPIN3NPA subunit A structures in different orientations. The r.m.s.d. for 377 Cα atoms in two structures is 0.34 Å. c, d, The inverted structural repeat of 5 TMs in TMD of AtPIN3apo. c, Structures of TM1–5 (blue) and TM6–10 (cyan) shown together or individually in the same orientation. d, Structural superimposition of TM1–5 and TM6–10 in different orientations.
Extended Data Fig. 6 Structure and topology comparisons of AtPIN3, SLC9A, and SLC10A transporters.
The structure of each transporter is shown in the side (left) and top (middle) views. The topology (right) is presented with the extracellular side on the top and the intracellular side at the bottom. a, AtPIN3; b, ASBTYf, Yersinia frederiksenii ASBT, PDB: 4N7W; c, ASBTNm, Neisseria meningitidis ASBT, PDB: 3ZUY; d, HsNHE1, human NHE1, PDB: 7DSW; e, TtNapA, Thermus thermophilus NapA, PDB: 5BZ2; f, MjNhaP, Methanocaldococcus jannaschii NhaP, PDB: 4CZB.
Extended Data Fig. 7 Ligand-binding sites in AtPIN3.
a, The NPA-bound structure of AtPIN3NPA (left), the taurocholic acid (TCA)-bound structure of ASBTYf (middle), and the modelled IAA-bound structure of AtPIN3IAA (right). NPA, TCA, and IAA are shown in cyan, green, and yellow, respectively. b, c, The RMSD of AtPIN3 (b) and IAA (c) during the 400 ns simulation for four trajectories. d–g, The binding configurations of IAA in four trajectories extracted from every 20 ns of simulation (green, cyan, purple, and orange) is similar to that of modelled IAA in the AtPIN3IAA structure (yellow). h, Time series showing the shortest observed distance among AtPIN3 residues to IAA. Numbers in parentheses are the mean and standard deviation of respective distances in MD simulations. i, Snapshots of IAA binding site for one of the trajectories, taken every 80 ns and spanning 400 ns. IAA in the AtPIN3IAA structure is in yellow and in MD simulations in green; AtPIN3 in the structure of AtPIN3IAA is in magenta and in MD simulations in pink.
Extended Data Fig. 8 SPR assay of AtPIN3 mutants with IAA or NPA.
a, SPR binding curves for the interaction of AtPIN3 mutants with IAA or NPA at different concentrations. b, KD curves of AtPIN3 and IAA (left) or NPA (right) fitted with a steady-state affinity model. c, The KD values of IAA or NPA bound to WT and mutant AtPIN3 calculated using the SPR steady-state affinity model. d, SPR binding curves for the interaction of WT AtPIN3 with IAA at different pH conditions. e, KD curves of WT AtPIN3 and IAA at different pH conditions fitted with a steady-state affinity model.
Extended Data Fig. 9 Dimeric assembly of AtPIN3 and SLC9A transporters in inward-facing conformations.
a, Localization of residues for mutagenesis in AtPIN3apo. b and c, Size-exclusion chromatography of AtPIN3 with mutations at the dimer interface of scaffold domain. d, The native PAGE of WT and two quadruple mutants. n = 3 independent experiments. e, The 3H-IAA efflux of WT and mutant AtPIN3 at 10 min in the IAA loading phase (n = 6 independent experiments). Data were presented as mean ± s.e.m. Two-tailed t-test was performed. f–i, Dimeric structures of AtPIN3apo (f), HsNHE1 (g, PDB: 7DSW), TtNapA (h, PDB: 5BZ2), and MjNhaP (i, PDB: 4CZB) in extracellular view (left), side view (middle), and cut-in surface model (right). The intracellular-facing cavity and extracellular-facing cavity of AtPIN3 are indicated by arrows.
Supplementary information
Supplementary Figure 1
Uncropped images. a, Uncropped blots for Extended Data Fig. 2c. b, Uncropped gels for Extended Data Fig. 3a,f,k. c, Uncropped gel for Extended Data Fig. 9d.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, N., Zhu, A., Tao, X. et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature 609, 616–621 (2022). https://doi.org/10.1038/s41586-022-05142-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05142-w
This article is cited by
-
3,4-Dichlorophenylacetic acid acts as an auxin analog and induces beneficial effects in various crops
Communications Biology (2024)
-
Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1
Nature Plants (2024)
-
Design and synthesis of strong root gravitropism inhibitors with no concomitant growth inhibition
Scientific Reports (2023)
-
Molecular mechanisms of N-1-naphthylphthalamic acid, a chemical tool in plant biology and agriculture
Molecular Horticulture (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.