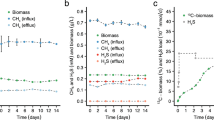
Abstract
Methane (CH4), the most abundant hydrocarbon in the atmosphere, originates largely from biogenic sources1 linked to an increasing number of organisms occurring in oxic and anoxic environments. Traditionally, biogenic CH4 has been regarded as the final product of anoxic decomposition of organic matter by methanogenic archaea. However, plants2,3, fungi4, algae5 and cyanobacteria6 can produce CH4 in the presence of oxygen. Although methanogens are known to produce CH4 enzymatically during anaerobic energy metabolism7, the requirements and pathways for CH4 production by non-methanogenic cells are poorly understood. Here, we demonstrate that CH4 formation by Bacillus subtilis and Escherichia coli is triggered by free iron and reactive oxygen species (ROS), which are generated by metabolic activity and enhanced by oxidative stress. ROS-induced methyl radicals, which are derived from organic compounds containing sulfur- or nitrogen-bonded methyl groups, are key intermediates that ultimately lead to CH4 production. We further show CH4 production by many other model organisms from the Bacteria, Archaea and Eukarya domains, including in several human cell lines. All these organisms respond to inducers of oxidative stress by enhanced CH4 formation. Our results imply that all living cells probably possess a common mechanism of CH4 formation that is based on interactions among ROS, iron and methyl donors, opening new perspectives for understanding biochemical CH4 formation and cycling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data reported in this paper are available in the Supplementary Information provided with this paper and on request from the corresponding authors. Source data are provided with this paper.
References
Saunois, M. et al. The global methane budget 2000–2017. Earth Syst. Sci. Data 12, 1561–1623 (2020).
Keppler, F., Hamilton, J. T. G., Braß, M. & Röckmann, T. Methane emissions from terrestrial plants under aerobic conditions. Nature 439, 187–191 (2006).
McLeod, A. R. et al. Ultraviolet radiation drives methane emissions from terrestrial plant pectins. New Phytol. 180, 124–132 (2008).
Lenhart, K. et al. Evidence for methane production by saprotrophic fungi. Nat. Commun. 3, 1046 (2012).
Klintzsch, T. et al. Methane production by three widespread marine phytoplankton species: release rates, precursor compounds, and potential relevance for the environment. Biogeosciences 16, 4129–4144 (2019).
Bižić, M. et al. Aquatic and terrestrial cyanobacteria produce methane. Sci. Adv. 6, eaax5343 (2020).
Thauer, R. K. Methyl (alkylalkyl)-coenzyme M reductases: nickel F-430-containing enzymes involved in anaerobic methane formation and in anaerobic oxidation of methane or of short chain alkanes. Biochemistry 58, 5198–5220 (2019).
Conrad, R. The global methane cycle: recent advances in understanding the microbial processes involved. Env. Microbiol. Rep. 1, 285–292 (2009).
DeLong, E. F. Exploring marine planktonic archaea: then and now. Front. Microbiol. 11, 3527 (2021).
Vorholt, J., Kunow, J., Stetter, K. O. & Thauer, R. K. Enzymes and coenzymes of the carbon monoxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus. Arch. Microbiol. 163, 112–118 (1995).
Hartmann, J. F. et al. High spatiotemporal dynamics of methane production and emission in oxic surface water. Environ. Sci. Technol. 54, 1451–1463 (2020).
Kamat, S. S., Williams, H. J., Dangott, L. J., Chakrabarti, M. & Raushel, F. M. The catalytic mechanism for aerobic formation of methane by bacteria. Nature 497, 132–136 (2013).
Metcalf, W. W. et al. Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science 337, 1104–1107 (2012).
Zheng, Y. et al. A pathway for biological methane production using bacterial iron-only nitrogenase. Nat. Microbiol. 3, 281–286 (2018).
North, J. A. et al. A nitrogenase-like enzyme system catalyzes methionine, ethylene, and methane biogenesis. Science 369, 1094–1098 (2020).
Wang, Q. et al. Aerobic bacterial methane synthesis. Proc. Natl Acad. Sci. USA 118, e2019229118 (2021).
Postgate, J. R. Methane as a minor product of pyruvate metabolism by sulphate-reducing and other bacteria. J. Gen. Microbiol. 57, 293–302 (1969).
Althoff, F. et al. Abiotic methanogenesis from organosulphur compounds under ambient conditions. Nat. Commun. 5, 4205 (2014).
Enami, S., Sakamoto, Y. & Colussi, A. J. Fenton chemistry at aqueous interfaces. Proc. Natl Acad. Sci. USA 111, 623–628 (2014).
Mittler, R. ROS are good. Trends Plant Sci. 22, 11–19 (2017).
Braun, V. & Hantke, K. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 15, 328–334 (2011).
Dunbar, K. L., Scharf, D. H., Litomska, A. & Hertweck, C. Enzymatic carbon–sulfur bond formation in natural product biosynthesis. Chem. Rev. 117, 5521–5577 (2017).
Wax, R. & Freese, E. Initiation of the germination of Bacillus subtilis spores by a combination of compounds in place of l-alanine. J. Bacteriol. 95, 433–438 (1968).
Ewing, D. The effects of dimethylsulfoxide (DMSO) on the radiation sensitivity of bacterial spores. Radiat. Res. 90, 348–355 (1982).
Setlow, B., Melly, E. & Setlow, P. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J. Bacteriol. 183, 4894–4899 (2001).
Candeias, L. P., Stratford, M. R. L. & Wardman, P. Formation of hydroxyl radicals on reaction of hypochlorous acid with ferrocyanide, a model iron(II) complex. Free Radical Res. 20, 241–249 (2009).
Bruskov, V. I., Masalimov, Z. K. & Chernikov, A. V. Heat-induced generation of reactive oxygen species in water. Dokl. Biochem. Biophys. 384, 181–184 (2002).
Foyer, C. H. & Noctor, G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18 (2011).
Rush, J. D. & Koppenol, W. H. Reactions of Fe(II)–ATP and Fe(II)–citrate complexes with t-butyl hydroperoxide and cumyl hydroperoxide. FEBS Lett. 275, 114–116 (1990).
Wongnate, T. et al. The radical mechanism of biological methane synthesis by methyl-coenzyme M reductase. Science 352, 953–958 (2016).
Ross, M. O. & Rosenzweig, A. C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 22, 307–319 (2016).
Mols, M. & Abee, T. Primary and secondary oxidative stress in Bacillus. Environ. Microbiol. 13, 1387–1394 (2011).
Wishkerman, A. et al. Enhanced formation of methane in plant cell cultures by inhibition of cytochrome c oxidase. Plant Cell Environ. 34, 457–464 (2011).
Tuboly, E. et al. Methane biogenesis during sodium azide-induced chemical hypoxia in rats. Am. J. Physiol. Cell Physiol. 304, 207–214 (2013).
Klintzsch, T. et al. Effects of temperature and light on methane production of widespread marine phytoplankton. J. Geophys. Res. Biogeosci. 125, e2020JG005793 (2020).
Polag, D., Leiß, O. & Keppler, F. Age dependent breath methane in the German population. Sci. Total Environ. 481, 582–587 (2014).
Zhang, X. et al. Methane limit LPS-induced NF-κB/MAPKs signal in macrophages and suppress immune response in mice by enhancing PI3K/AKT/GSK-3β-mediated IL-10 expression. Sci. Rep. 6, 293591 (2016).
Qaderi, M. M. & Reid, D. M. Methane emissions from six crop species exposed to three components of global climate change: temperature, ultraviolet-B radiation and water stress. Physiol. Plant 137, 139–147 (2009).
Brüggemann, N. et al. Nonmicrobial aerobic methane emission from poplar shoot cultures under low-light conditions. New Phytol. 182, 912–918 (2009).
Harwood, C. R. & Cutting, S. M. (eds) Molecular Biological Methods for Bacillus. Vol. 1 (John Wiley & Sons, 1990).
Mutlu, A. et al. Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity–quality tradeoff. Nat. Commun. 9, 69 (2018).
Acknowledgements
We thank M. Schneider, M. Schroll, J. Hädeler, I. Ralenekova, M. Greule, B. Knape, S. Rheinberger, C. Kaspar, Y. Gietz, C. Walter, E. Wiedtke, N. Fischer and L. Dietz for technical support; T. Erb, S. Greiner, M. Baumgart, M. Knop and R. Fischer for providing strains (affiliations included in the Supplementary information); R. Thauer for providing critical comments; and P. Hardy and J. Hamilton for editing the manuscript. Figures 1, 2a and 3b,c, Extended Data Fig. 6 and Supplementary Fig. 7a were created with BioRender.com. This work was supported by funds from the Max Planck Society (I.B.B., J.G.R., L.E., B.S.), Friedrich Naumann Foundation (L.E.) and the Landesgraduiertenstiftung Baden-Württemberg (B.S.). L.E., J.G.R., F.K., T.K. and D.G. were supported by the German Research Foundation (DFG; 446841743, KE 884/8-2 and KE 884/16-2). The EPR core facility was supported by 3DMM2O–Cluster of Excellence (EXC-2082/1–390761711) at Heidelberg University.
Author information
Authors and Affiliations
Contributions
L.E., I.B.B. and F.K. designed the research. L.E. performed the experiments. U.B., B.S. and M.K. contributed to the experiments and analysed the following data: Fig. 2c, Extended Data Fig. 4 and Supplementary Figs. 5 and 9 (U.B.); Extended Data Fig. 1 (B.S.); and Supplementary Fig. 5 (M.K.). L.E., B.S., U.B., T.K., J.G.R., I.B.B. and F.K analysed all other data. I.B.B., T.P.D., D.G., J.G.R. and F.K. supervised the research. L.E., I.B.B. and F.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Stephen Ragsdale, Marc Strous, David Valentine and Jingyao Zhang for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Germinated, but not dormant, B. subtilis spores generate CH4 from 2H-DMSO.
Scatterplot depicting 2H % excess and amounts of CH4 formed. The second y-axis shows the calculated corresponding fraction of methyl precursor compound (FMPC [%]) involved in CH4 generation. Insets show brightfield micrographs of refractile dormant spores and non-refractile germinated spores (see Methods). DMSO was 95 % unlabeled (~natural abundance) and 5 % 2H labelled. A 2H % excess of 3.75 % correlates with 100 % FMPC (as CH4 is formed from a 2H-labelled methyl group, containing three hydrogen atoms, and a fourth, unlabelled hydrogen atom). Amounts of CH4 emitted were calculated by subtracting the media controls from the spore cultures. Germination was induced by adding 3 mL of AGFK (19.8 mM L-asparagine, 33.6 mM D-glucose, 33.6 mM D-fructose and 60 mM KCl). Data points represent individual measurements from N = 3 technical replicates from one experiment.
Extended Data Fig. 2 Methane formation and population growth of B. subtilis correlate with supply of fresh air.
While population growth (brown line) and CH4 formation (yellow bars) stop under anoxic conditions, both are initiated again upon supply of fresh air (air: ~21 % O2 and 78 % N2). Here, B. subtilis was grown at 37 °C in LB medium (starting OD600nm = 0.12), supplemented with 200 mM DMSO, in atmospherically-sealed glass vials with a volume of 20 mL including 10 mL bacterial culture and 10 mL headspace. At 1 h incubation intervals, the headspace gas was sampled and analysed. Anoxic conditions were generated by exchanging the headspace gas with a nitrogen atmosphere after drawing vacuum four times. Oxic conditions were restored by removing the vial seals for 3 min allowing entry of atmospheric air. Amounts of CH4 emitted were calculated by subtracting atmospheric background CH4 levels from measured CH4 levels. Six rounds of anoxia-reoxygenation were performed in order to establish that the process could occur repeatedly. Data points represent individual measurements, bars represent means from N = 2 technical replicates as guide to the eye.
Extended Data Fig. 3 Methane formation by B. subtilis is enhanced by oxidative and environmental stressors.
In comparison to unstressed cultures, CH4 levels (yellow) increase significantly upon treatment with oxidants HOCl or H2O2. Environmental stressors NaCl (salt stress) and heat (50 °C) also significantly enhance CH4 formation by B. subtilis. Pre-cultures were grown at 37 °C in LB media (starting OD600nm = 0.01) for 10 h and subsequently supplemented with 100 mM DMSO and, optionally, 300 µg mL−1 HOCl, 1 mM H2O2 or 4 % NaCl. 30 mL cultures and corresponding media controls were incubated at 37 °C or 50 °C (heat stress) in 60 mL sealed glass vials for 8 h. Amounts of CH4 emitted were calculated by subtracting media controls from culture samples. Bars represent means ± SD from N = 3 technical replicates from one experiment, respectively. Statistical analysis was performed using paired two-tailed t-tests, ***: p ≤ 0.001. Data points represent individual measurements.
Extended Data Fig. 4 B. subtilis ROS levels are promoted by oxidative stress and reduced by antioxidants.
In B. subtilis cultures treated with iron (F) and DMSO (S), bacterial ROS levels are enhanced upon oxidative stress induction (O) and reduced upon antioxidant addition. B. subtilis pre-cultures were grown in S750 media (starting OD600nm = 0.01), containing 50 nM FeSO4 and, optionally, 500 µM butylated hydroxytoluene (A) or Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) acting as assay validation antioxidant (V) for 6 h under standard conditions (37 °C, 180 rpm). Subsequently, cultures were supplemented with 4.95 µM FeSO4 and 200 mM DMSO and grown for 2 h. Cultures were next diluted 10-fold, supplemented with 25 µM 5-(and-6)-chloromethyl 2ʹ,7ʹ-dichlorofluorescin diacetate (CM-H2DCFDA) and incubated for 30 min in the dark. Finally, oxidative stress was induced by adding 150 µg mL−1 HOCl. After 10 min incubation, DCF fluorescence was measured from live cell population with FACS Canto II (BD) at green channel (excitation: 480 nm, emission: 520 nm) and analyzed with analyzed using FlowJo™ v10.8 Software (BD Life Sciences). (A). Gating strategy for flow cytometry. For all samples, the identical gating strategy was applied. (B) The assay was validated by supplying cultures with CM-H2DCFDA, HOCl and Trolox. (C) Detected cellular fluorescence distributions with and without BHT treatment, indicating respective ROS levels of cells stressed with HOCl. (D) Median values indicating the different cellular ROS levels from the obtained distributions. Numbers in brackets denote the respective analyzed cell counts.
Extended Data Fig. 5 B. subtilis forms CH4 from methylated S-/N-compounds.
2H % excess, the corresponding fraction methyl precursor compound (FMPC [%]) and amounts of CH4 produced in the presence of the indicated substrates provide unambiguous evidence for CH4 formation from methionine (yellow), trimethylamine (green) and DMSO (red). In contrast, no significant CH4 formation from the quaternary compound choline (blue) was detected. In comparison to cultures grown under standard conditions (circles), all of the three measured readouts increased in stressed cultures (squares). All examined compounds except DMSO were 95 % unlabelled and 5 % 2H-labelled. Among the 5 % labelled compounds, all methyl groups were fully 2H-labelled, implying that a 2H % excess of 3.75 % correlates with 100 % FMPC (as CH4 is formed from a 2H-labelled methyl group, containing three hydrogen atoms, and a fourth, unlabelled hydrogen atom). As DMSO was 2 % labelled, a 2H % excess of 1.5 % correlates with 100 % FMPC. The reason for addition of only 2 % labelled DMSO was due to the large amounts of CH4 formed and practical reasons for the IRMS measurements (limitations of measuring CH4 highly enriched in 2H). A pre-culture (Start OD600nm = 0.01) was grown in S750 medium, supplemented with 50 µM FeCl3, incubated for 8 h at 37 °C and 180 rpm. The pre-culture was subsequently split into 30 mL fractions. Both these fractions and media controls were incubated in sealed 60 mL glass vials for 24 h under identical conditions and supplemented with 50 mM substrate and, optionally, 300 µg mL−1 HOCl in order to induce oxidative stress. Methane amounts were measured by GC-FID and isotope values by GC-TC-IRMS (see SI Methods section). CH4 emission was calculated by subtracting media controls from corresponding bacterial cultures. It is proposed that methionine, trimethylamine and DMSO serve as substrates for ROS-driven CH4 formation (see Extended Data Fig. 6).
Extended Data Fig. 6 Model of ROS-driven CH4 formation from endogenous substrates.
Metabolic pathways and biochemical C1-transfer exist in a variety of different organisms and facilitate the production of substrates suitable for ROS-driven CH4 formation from endogenous precursor compounds. The methyl group of pyruvate (labelled in red) can be transferred to substrates for ROS-driven CH4 formation, e.g. methionine, or natural products, e.g. DMSO or TMA, which are produced by several organisms.
Extended Data Fig. 7 Methane formation in vitro is driven by H2O2 and enhanced by Fe3+-reductants and Fenton-activating Fe2+-chelators.
S750 media (grey), supplemented with 50 µM FeCl3 and 200 mM DMSO, was treated with 50 mM H2O2 (dashed lines), 1 mM ascorbate (red), glutathione (blue) or NADH (green) and 2 mM ATP, citrate or EDTA and incubated for 24 h at a neutral pH, 37 °C and 180 rpm. Upon H2O2 supplementation, CH4 formation in the non-supplemented media increased by ~19-fold. Addition of Fe3+-reductants ascorbate, glutathione or NADH enhanced CH4 formation by factors of ~83, ~13 or ~2, respectively. Upon addition of Fenton-activating Fe2+-chelators ATP, citrate or EDTA, the observed CH4 formation in media supplemented with H2O2 and ascorbate further increased by factors of ~8 (ATP), ~2 (citrate) or ~22 (EDTA). In media supplemented with H2O2 and glutathione, CH4 formation increased by factors of ~7 (ATP), ~3 (citrate) or ~26 (EDTA). In media supplemented with H2O2 and NADH, CH4 formation increased by factors of ~5 (ATP), ~4 (citrate) or ~19 (EDTA). The dashed red line shows background (laboratory air) CH4 content. Sealed vials with a volume of 40 mL including 20 mL sample and 20 mL headspace were used for incubations. Data points represent individual measurements, bars represent means from N = 2 technical replicates as guide to the eye.
Extended Data Fig. 8 Bacteria facilitate ROS-driven CH4 formation.
Overall, of the 19 different species of bacteria investigated all form CH4 (yellow) which is enhanced upon HOCl-induced oxidative stress induction (yellow, dashed lines). Bacteria were cultivated in Terrific Broth (TB) medium, supplemented with 1 % Glucose, 0.1 % K-Glutamate, 50 µg mL−1 L-Tryptophan, 50 µM MnCl2, 5 µM FeSO4, 2 mM MgSO4, 2 µM Thiamine, 1 µM ZnCl2, 0.7 mM CaCl2, 1.5 mM NaCl, 100 mM DMSO and, optionally, 150 µg mL−1 HOCl. Furthermore, Thermus thermophilus HB27 and Halomonas GFAJ-1 were supplemented with additional 30 mM and 500 mM NaCl, respectively. Stationary-phase bacterial pre-cultures (100 µL) were added to 9.9 mL media in 20 mL vials and incubated for 24 h at 37 °C with shaking at 180 r.p.m. For statistical analysis, averages for each duplicate were calculated and a two-tailed Wilcoxon signed rank test was performed (H0 = no difference between stressed and unstressed cultures). Thus, the observed differences between stressed and unstressed cultures could be demonstrated to be significant (p ≤ 0.001). Data points represent individual measurements, bars represent means from N = 2 technical replicates as guide to the eye.
Extended Data Fig. 9 Human HEK293T cells convert 2H-DMSO into CH4.
2H % excess, the corresponding fraction methyl precursor compound (FMPC [%]) and amounts of CH4 produced in the presence of 2H-DMSO provide unambiguous evidence for CH4 formation by HEK293T cells. 30 mL HEK293T cells (starting from 6*105 cells mL−1) and media controls were grown in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10 % Fetal Calf Serum (FBS), 50 nM FeSO4 and 50 mM DMSO and incubated at 37 °C with shaking at 50 r.p.m. for 48 h in 60 mL atmospherically-sealed glass vials. DMSO was 95 % unlabelled and 5 % labelled. Among the 5 % 2H-DMSO, all methyl groups were fully 2H-labelled, implying that a 2H % excess of 3.75 % correlates with 100 % FMPC (as CH4 is formed from a 2H-labelled methyl group, containing three hydrogen atoms, and a fourth, unlabelled hydrogen atom). CH4 emission was calculated by subtracting media control from cell culture amounts. Methane amounts were measured by GC-FID and isotope values determined by GC-TC-IRMS (see Methods section).
Extended Data Fig. 10 ROS-driven CH4 formation by mammalian cell lines.
30 mL HEK293T, HeLa and A549 cells (starting from 6*105 cells mL−1) and media controls were grown in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10 % Fetal Calf Serum (FBS) and 50 mM DMSO at 37 °C with shaking at 50 rpm for 96 h in 60 mL atmospherically-sealed glass vials. HL60 cells (starting from 1*106 cells mL−1) were grown in RPMI medium with identical supplementations and incubation conditions. Optionally, oxidative stress conditions were induced by supplementing cultures and media controls with 150 µg mL−1 HOCl (orange, dashed lines). Amounts of CH4 emitted were calculated by subtracting media controls from respective cell culture samples. Bars represent means ± SD from N = 3 technical replicates from one experiment. Statistical analysis was performed using paired two-tailed t-tests, **: p ≤ 0.01, ***: p ≤ 0.001. Data points represent individual measurements.
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–10, a list of the organisms used and the Supplementary References.
Supplementary Data
Data underlying Supplementary Figs. 1–10.
Source data
Rights and permissions
About this article
Cite this article
Ernst, L., Steinfeld, B., Barayeu, U. et al. Methane formation driven by reactive oxygen species across all living organisms. Nature 603, 482–487 (2022). https://doi.org/10.1038/s41586-022-04511-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04511-9
This article is cited by
-
Optimization strategy of Co3O4 nanoparticles in biomethane production from seaweeds and its potential role in direct electron transfer and reactive oxygen species formation
Scientific Reports (2024)
-
Floating macrophyte phyllosphere as a habitat for methanogens
Environmental Chemistry Letters (2024)
-
Characterization and environmental applications of soil biofilms: a review
Environmental Chemistry Letters (2024)
-
Phosphonate consumers potentially contributing to methane production in Brazilian soda lakes
Extremophiles (2024)
-
Methane formation driven by light and heat prior to the origin of life and beyond
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.