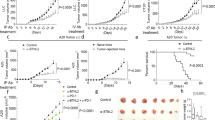
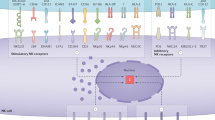
Abstract
Cytokines were the first modern immunotherapies to produce durable responses in patients with advanced cancer, but they have only modest efficacy and limited tolerability1,2. In an effort to identify alternative cytokine pathways for immunotherapy, we found that components of the interleukin-18 (IL-18) pathway are upregulated on tumour-infiltrating lymphocytes, suggesting that IL-18 therapy could enhance anti-tumour immunity. However, recombinant IL-18 previously did not demonstrate efficacy in clinical trials3. Here we show that IL-18BP, a high-affinity IL-18 decoy receptor, is frequently upregulated in diverse human and mouse tumours and limits the anti-tumour activity of IL-18 in mice. Using directed evolution, we engineered a ‘decoy-resistant’ IL-18 (DR-18) that maintains signalling potential but is impervious to inhibition by IL-18BP. Unlike wild-type IL-18, DR-18 exerted potent anti-tumour effects in mouse tumour models by promoting the development of poly-functional effector CD8+ T cells, decreasing the prevalence of exhausted CD8+ T cells that express the transcriptional regulator of exhaustion TOX, and expanding the pool of stem-like TCF1+ precursor CD8+ T cells. DR-18 also enhanced the activity and maturation of natural killer cells to effectively treat anti-PD-1 resistant tumours that have lost surface expression of major histocompatibility complex class I molecules. These results highlight the potential of the IL-18 pathway for immunotherapeutic intervention and implicate IL-18BP as a major therapeutic barrier.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated during this study are available within the paper. The scRNA-seq data were deposited on Gene Expression Omnibus (GSE146609). Source data are provided with this paper.
References
Atkins, M. B., Kunkel, L., Sznol, M. & Rosenberg, S. A. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J. Sci. Am. 6 (Suppl. 1), S11–S14 (2000).
Waldmann, T. A. Cytokines in cancer immunotherapy. Cold Spring Harb. Perspect. Biol. 10, a028472 (2018).
Tarhini, A. A. et al. A phase 2, randomized study of SB-485232, rhIL-18, in patients with previously untreated metastatic melanoma. Cancer 115, 859–868 (2009).
Spangler, J. B., Moraga, I., Mendoza, J. L. & Garcia, K. C. Insights into cytokine-receptor interactions from cytokine engineering. Annu. Rev. Immunol. 33, 139–167 (2015).
Mantovani, A., Dinarello, C. A., Molgora, M. & Garlanda, C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 50, 778–795 (2019).
Guo, L., Junttila, I. S. & Paul, W. E. Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol. 33, 598–606 (2012).
Ma, Z. et al. Augmentation of immune checkpoint cancer immunotherapy with IL18. Clin. Cancer Res. 22, 2969–2980 (2016).
Hu, B. et al. Augmentation of antitumor immunity by human and mouse CAR T cells secreting IL-18. Cell Rep. 20, 3025–3033 (2017).
Robertson, M. J. et al. Clinical and biological effects of recombinant human interleukin-18 administered by intravenous infusion to patients with advanced cancer. Clin. Cancer Res. 12, 4265–4273 (2006).
Dinarello, C. A., Novick, D., Kim, S. & Kaplanski, G. Interleukin-18 and IL-18 binding protein. Front. Immunol. 4, 289 (2013).
Robertson, M. J. et al. A dose-escalation study of recombinant human interleukin-18 using two different schedules of administration in patients with cancer. Clin. Cancer Res. 14, 3462–3469 (2008).
Kim, S. H. et al. Site-specific mutations in the mature form of human IL-18 with enhanced biological activity and decreased neutralization by IL-18 binding protein. Proc. Natl Acad. Sci. USA 98, 3304–3309 (2001).
Simoni, Y. et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018).
Salmon, H. et al. Expansion and activation of CD103+ dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 44, 924–938 (2016).
Apetoh, L. et al. Consensus nomenclature for CD8+ T cell phenotypes in cancer. OncoImmunology 4, e998538 (2015).
Best, J. A. et al. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412 (2013).
Wherry, E. J. & Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1−CD8+ tumor-infiltrating T cells. Immunity 50, 181–194.e186 (2019).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Hudson, W. H. et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058.e1044 (2019).
Murray, P. J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Gubin, M. M. et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175, 1014–1030.e1019 (2018).
Jeannet, G. et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl Acad. Sci. USA 107, 9777–9782 (2010).
Gattinoni, L. et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 15, 808–813 (2009).
Sade-Feldman, M. et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 8, 1136 (2017).
Ardolino, M. et al. Cytokine therapy reverses NK cell anergy in MHC-deficient tumors. J. Clin. Invest. 124, 4781–4794 (2014).
Street, S. E., Trapani, J. A., MacGregor, D. & Smyth, M. J. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J. Exp. Med. 196, 129–134 (2002).
Nakamura, K. et al. Dysregulated IL-18 is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell 33, 634–648.e635 (2018).
Terme, M. et al. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res. 71, 5393–5399 (2011).
Vidal-Vanaclocha, F. et al. IL-18 regulates IL-1beta-dependent hepatic melanoma metastasis via vascular cell adhesion molecule-1. Proc. Natl Acad. Sci. USA 97, 734–739 (2000).
Park, C. C. et al. Evidence of IL-18 as a novel angiogenic mediator. J. Immunol. 167, 1644–1653 (2001).
Hirakawa, M. et al. Low-dose IL-2 selectively activates subsets of CD4+ Tregs and NK cells. JCI Insight 1, e89278 (2016).
Rosenberg, S. A. IL-2: the first effective immunotherapy for human cancer. J. Immunol. 192, 5451–5458 (2014).
Belkaya, S. et al. Inherited IL-18BP deficiency in human fulminant viral hepatitis. J. Exp. Med. 216, 1777–1790 (2019).
Wang, J. et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30, 428–435 (2017).
Carbotti, G. et al. The IL-18 antagonist IL-18-binding protein is produced in the human ovarian cancer microenvironment. Clin. Cancer Res. 19, 4611–4620 (2013).
Lin, L. et al. Interleukin-37 expression and its potential role in oral leukoplakia and oral squamous cell carcinoma. Sci. Rep. 6, 26757 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Heng, T. S. & Painter, M. W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Singer, M. et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell 166, 1500–1511.e1509 (2016).
Acknowledgements
We thank all members of the Ring and Bosenberg labs for advice and technical assistance; E. Folta-Stogniew for assistance with SPR; and A. Iwasaki for mice and reagents. The cartoon in Fig. 1a was created using BioRender (https://biorender.com/). This work was supported by grants from the National Cancer Institute Immuno-Oncology Translation Network (U01CA233096; to A.M.R. and M.W.B.), Gabrielle’s Angel Foundation (to A.M.R.), and the Blavatnik Fund for Innovation at Yale (to A.M.R.). A.M.R. is additionally supported by an NIH Director’s Early Independence Award (DP5OD023088), the Pew-Stewart Scholars Program, and the Robert T. McCluskey Foundation. M.F.S. is supported by a Miguel Servet contract from Instituto de Salud Carlos III, Fondo de Investigacion Sanitaria (Spain). W.D. is supported by a Career Development Award from the Dermatology Foundation. O.-E.W. is supported by a NSF Graduate Research Fellowship (1752134) and by a NIH training grant (T32AI055403). The T100 Biacore instrumentation used was supported by NIH Award S10RR026992-0110.
Author information
Authors and Affiliations
Contributions
T.Z., W.D., O.-E.W., K.P.H., M.W.B., and A.M.R. designed experiments. T.Z., W.D., O.-E.W., K.P.H., C.E.R., and S.F. performed experiments. T.Z., W.D., O.-E.W., M.K.M., K.P.H., J.W., M.W.B., and A.M.R. analysed data. M.F.S. provided samples from patients with NSCLC. R.J. and R.A.F. provided Il18bp−/− and Il18r1−/− mice. T.Z., O.-E.W., W.D., M.W.B., and A.M.R. wrote the paper. M.W.B. and A.M.R. supervised the research. A.M.R. conceived of the project.
Corresponding author
Ethics declarations
Competing interests
A.M.R., T.Z., and S.F. are named inventors on a patent application that describes the DR-18 molecule. A.M.R. is the founder of Simcha Therapeutics, the commercial licensee of DR-18, and holds equity in the immuno-oncology companies Forty-Seven Inc., ALX Oncology, and Medicenna Therapeutics. W.D. and M.W.B. serve as consultants for Eli Lilly. W.D. has research funding from Pfizer for unrelated work.
Additional information
Peer review information Nature thanks Jim Wells, Dietmar Zehn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Regulation of the IL-18 signalling axis in the TME.
a, Distribution of intratumoral CD8+ T cell transcripts by dysfunction and activation gene scores, adapted from Singer et al.43. Blue points mark a curated list of cytokine and cytokine receptors. Red points mark Il18, Il18r1, Il18rap. b–f, Wild-type C57BL/6 mice were subcutaneously engrafted with the indicated mouse syngeneic tumour. Tumour and spleen were removed and analysed by flow cytometry at day 7 after engraftment. b, c, Representative flow plots (b) and quantification of percentage of IL-18Rα expression (c) on splenic and intratumoral CD4+ T cells (NK1.1−, CD3+, TCRβ+, CD4+), CD8+ T cells (NK1.1−, CD3+, TCRβ+, CD8+), NK cells (CD3−, TCRβ−, NK1.1+, CD200R1−) from wild-type mice engrafted with MC38 cells. d, Percentage IL-18Rα expression on splenic and intratumoral CD4+ T cells, CD8+ T cells, and NK cells from wild-type mice engrafted with YUMMER1.7 cells. e, f, Representative flow plots (e) and quantification of surface CD44 and IL-18Rα expression (f) on CD8+ T cells in spleen and tumour from MC38 tumour-bearing mice. g, Normalized quantification of Il18bp by qPCR in blood and tumour lysate assessed at day 7 after s.c. engraftment of wild-type mice with MC38 tumours, treated with PBS, IL-18 or anti-IFN-γ + IL-18. h, Quantification of plasma IL-18BP protein level by ELISA at day 7 after s.c. engraftment of wild-type mice with MC38 tumours, treated with either PBS or IL-18. i, Representative immunohistochemistical staining for IL-18BP from l18bp−/− mouse spleen, wild-type spleen, MC38 tumour, or MC38 tumour treated with one dose of IL-18, and assessed on day 7. Scale bar, 25 μm. j, Quantification of IL-18BP+ cells per high power field (HPF) in representative sections from each group in i. b–f, i, Data are representative of two independent experiments with n = 5 mice per group. g, h, j, Data are pooled from two independent experiments with n = 5 mice per group. P values were calculated using two-way ANOVA (c, d, f) or two-tailed unpaired Student’s t-tests (g, h, j), and data are presented as mean ± s.e.m.
Extended Data Fig. 2 IL-18BP level is elevated in human cancers and correlated with T cell infiltration.
a, Expression of IL18BP transcripts in normal (blue) or cancer (red) tissues from the TCGA database. CHOL, cholangiocarcinoma; DLBC, diffuse large B cell lymphoma; GBM, glioblastoma multiforme; HSNC, head and neck squamous carcinoma; KIRC, kidney renal clear cell carcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma (*P < 0.01). b–d, Correlation of IL18BP expression with T cell markers CD3E (b), CD8A (c) and PDCD1 (d) from the TCGA database for SKCM (n = 558), BRCA (breast adenocarcinoma, n = 1,085), HNSC (n = 44), STAD (n = 221), and OV (ovarian cancer, n = 426). e, Frequency of IL-18BP immunohistochemistry staining levels in human tumour tissue microarrays. Each sample was scored as negative (0) or positive (1+, 2+, or 3+). Representative images are shown for each staining level. f, Quantification of plasma IL-18BP protein level by ELISA for healthy donors (n = 22) and patients with NSCLC (n = 52) at baseline before treatment and at the time of the following CT scan after receiving treatment with anti-PD-(L)1 (n = 52). g, Representative mean tumour growth in wild-type (left) and Il18bp−/− (right) mice engrafted s.c. with MC38 tumours and treated with PBS or IL-18. Data are representative of three independent experiments with n = 5 mice per group. P values were calculated using one-way ANOVA (a, f) or two-way ANOVA (g), and data are presented as mean ± s.e.m.
Extended Data Fig. 3 Generation of decoy-resistant IL-18.
Related to Fig. 1. a, Structural alignment of hIL-18 (green)–hIL-18Rα/Rβ (cyan) complex (PDB ID 3WO4) with hIL-18–vIL-18BP (blue) complex (PDB ID 3F62). b, c, Representative surface plasmon resonance (SPR) sensorgrams of mouse WT IL-18 binding to IL-18Rα (b) or IL-18BP (c). IL-18Rα measurements were conducted using a conventional multiple cycle program, whereas IL-18BP measurements were conducted using a single-cycle program. d, Dose–response curves of IL-18BP protein antagonizing IL-18Rα in complex with indicated IL-18 and mutants (E42A, K89A and E42A/K89A). Experiments were performed in duplicate (n = 2). e, Randomized positions of murine IL-18 to create DR_18, with the corresponding degenerate codon and the potential amino acid at each position. f, Summary of the experimental design for directed evolution and yeast selection process to generate DR-18. Yeast libraries were selected for IL-18Rα binding and counter-selected against IL-18BP using magnetic-activated cell sorting (MACS; rounds 1 and 2) and subsequently FACS (rounds 3–5). Blue text (right) indicates positive selection reagent, and red text (left) shows the counter-selection reagent. g, Structural representation of DR-18 mutation positions in IL-18Rα and IL-1BP binding overlap region. Side chains from a minimized set of mutations up to six consensus residues (1N, 50M, 52K, 55E, 56V and 59L) are displayed as stick models. b–d, Data are representative of two independent experiments and are presented as mean ± s.e.m.
Extended Data Fig. 4 Anti-tumour efficacy of DR-18 in syngeneic tumour models.
Related to Fig. 2. Wild-type mice were engrafted s.c. with 0.5 × 106 MC38 or YUMMER1.7 tumour cells. From day 7 after engraftment, when tumour size reached 50–100 mm3, mice were treated twice weekly with PBS, or 0.32 mg/kg IL-18 or DR-18. a, Representative spider plots depicting MC38 tumour growth in wild-type mice treated with PBS, IL-18, DR-18, anti-PD-1, IL-18 + anti-PD-1, or DR-18 + anti-PD-1. b, e, Representative mean tumour growth (a) and Kaplan–Meier survival curves (e) of wild-type mice engrafted s.c. with YUMMER1.7 cells and treated with PBS, IL-18, DR-18, anti-PD-1, IL-18 + anti-PD-1, or DR-18 + anti-PD-1. Arrows signify days of treatment. c, f, Mean tumour growth (c) and Kaplan–Meier survival curves (f) of wild-type Balb/c mice engrafted s.c. with 0.25 × 106 CT-26 colon carcinoma cells and treated with PBS, anti-PD-1, IL-18, or DR-18. d, g, Mean tumour growth (d) and and Kaplan–Meier survival curves (g) of wild-type mice engrafted s.c. with 0.25 × 106 B16-F10 melanoma cells and treated with PBS, IL-18, or DR-18 from day 7. h, Mean tumour growth of wild-type mice engrafted s.c. with MC38 cells and treated with PBS, IL-18, CS1 or CS2 (DR-18), with the indicated doses. a–d, h, Data are representative of three independent experiments with n = 5 mice per group. e–g, Data are summary of three (e) or two (f–h) independent experiments with n = 5 mice per group. Data are presented as mean ± s.e.m.
Extended Data Fig. 5 DR-18 acts on antigen-specific T cells in the tumour.
Related to Fig. 2. a, Kaplan–Meier survival curves of mice engrafted with YUMMER1.7 tumours and treated with PBS, DR-18, or DR-18 and anti-CD4, anti-CD8, anti-NK1.1, or anti-IFN-γ antibodies. Treatment parameters were the same as for MC38 tumours (Fig. 2a, see Methods). b, Kaplan–Meier survival curves of wild-type or Rag2−/− mice engrafted s.c. with MC38 tumours and treated with PBS or DR-18. c, Kaplan–Meier survival curves of mice re-challenged with 1.0 × 106 MC38 tumour cells after successful DR-18 treatment compared to naive mice engrafted with 1.0 × 106 MC38 cells for the first time. d–i, Thy1.1+ P14 CD8 T cells (2.0 × 106) were adoptively transferred into wild-type mice, which were then engrafted s.c. with 0.25 × 106 B16F10-gp33 tumour cells one day later. Mice were then treated twice weekly with PBS or DR-18 (0.32 mg/kg) once tumours reached 50–100 mm3 for a total of three treatments. d–f, Representative flow plots (d), quantification of frequency (e), and absolute number of antigen-specific tumour-infiltrating P14 CD8+ T cells (f), as defined by Thy1.1+ and Gp33-tetramer. g, Percentage of IFN-γ on intratumoral antigen-specific (Thy1.1+Tetramer+) P14 CD8+ cells after PBS (n = 4) or DR-18 (n = 4) treatment. h, i, Representative flow plots (h) and quantification (i) of CD44 and CD39 expression on endogenous (Thy1.1−Tetramer−) tumour-infiltrating CD8+ T cells. j, Quantification of CD44 and PD-1 expression on tumour-infiltrating CD8+ T cells from wild-type mice engrafted with MC38 cells and treated with PBS or DR-18 in combination with FTY720. k, Representative flow cytometry plots of IL-18Rα expression on CD4+ T cells (CD45+, NK1.1−, CD3+, TCRβ+, CD4+), CD8+ T cells (CD45+, NK1.1−, CD3+, TCRβ+, CD8+), NK cells (CD45+, CD3−, TCRβ−, NK1.1+), B cells (CD45+, CD3−, TCRβ−, NK1.1−, CD19+), monocytes (CD45+, CD64+, CD11b+, Ly6C+), macrophages (CD45+, CD64+, MHCII+, F4/80+), dendritic cells (CD45+, CD64−, CD11c+), and neutrophils (CD45+, CD64−, CD11b+, Ly6G+) from MC38 tumours (n = 5), matched spleens (n = 5), un-engrafted wild-type spleens (n = 3), or Il18r1−/− spleens (n = 3). l, Representative mean tumour growth of MC38 tumours implanted into Rag2−/− mice that were adoptively transferred with T cells isolated from wild-type (n = 5) or Il18r1−/− (n = 8) mice and treated with PBS or DR-18. Black arrows signify day of treatment. m, Representative mean tumour growth of MC38 tumours implanted into wild-type (n = 4) and XCR1DTR (n = 5) mice treated with PBS or DR-18 in combination with DT. Green arrows signify DT treatment regimen. d–j, Data are representative of two independent experiments and presented as mean ± s.e.m. P values were calculated using one-tailed (e, f) or two-tailed (g) unpaired Student’s t-tests or two-way ANOVA (i, j).
Extended Data Fig. 6 Single-cell transcriptomic landscape of the TME in DR-18-treated tumours.
Related to Fig. 3. Wild-type mice were engrafted s.c. with 0.5 × 106 YUMMER1.7 cells. From day 7 after engraftment, mice were treated twice weekly with PBS, IL-18 or DR-18 (s.c.). On day 15, tumours were removed and sorted, and 5,000 cells per condition were used for scRNA-seq analysis. a, t-SNE projection showing all cells present in YUMMER1.7 and YUMMER1.7-B2m−/− tumours, coloured by cluster. Cells from the YUMMER1.7-B2m−/− experiments are shown in light grey and are discussed later. b, t-SNE projection showing distribution of cells coloured by treatment condition (dark grey, PBS; blue, IL-18; magenta, DR-18). c, Percentage of cells in each cluster by treatment. d, t-SNE projection of all cells analysed showing expression of genes supporting cell-type assignments. e, t-SNE projections of all cells analysed showing expression of IL-18 pathway-related genes. f, t-SNE projections showing expression of selected genes in computationally isolated Cd3e-positive T cells. g, t-SNE projections of Cd68+ cells, showing expression of Cx3cr1, Nos2, Arg1, and Mertk. h, t-SNE projection of Cd68+ cells, coloured by graph-based cluster. i, t-SNE projection of myeloid cell distribution as a function of treatment condition. j, Frequency of macrophages within each cluster identified in h. k, Heatmap showing differentially expressed genes in Cd68+ cells by clusters identified in h. l, m, Frequency (l) and absolute numbers (m) of Nos2+ macrophages from MC38 engrafted tumours following PBS, IL-18 or DR-18 treatment. Data are representative of three independent experiments with n = 5 mice per group and presented as mean ± s.e.m. P values calculated using one-way ANOVA.
Extended Data Fig. 7 DR-18 treatment enhances lymphocyte effector function.
Related to Fig. 3. Wild-type mice were engrafted s.c. with 0.5 × 106 MC38 or YUMMER1.7 cells. From day 7 after engraftment mice were treated twice weekly i.p. with PBS, IL-18, or DR-18. Tumours were harvested and analysed on day 15 after engraftment. a, Absolute number of tumour-infiltrating CD4+ T cells, CD8+ T cells and NK cells from YUMMER1.7 tumours in each treatment group. b, Quantification of intracellular Ki-67+ staining by percentage of indicated intratumoral lymphocyte population. c, Absolute number of antigen-experienced (CD44+) CD8+ T cells from MC38 tumours in each treatment group. d, Representative flow plots of PD-1 and CD39 staining on intratumoral CD44+ CD8+ T cells from MC38 tumours in indicated treatment groups. e, Absolute number of PD-1−CD39−, PD-1+CD39− and PD-1+CD39+ CD44+CD8+ T cells from d in each treatment group. f, Quantification by Luminex of IFN-γ protein level from tumour lysate of indicated treatment groups. g, Quantification of intracellular IFN-γ staining by percentage of indicated intratumoral lymphocyte populations from YUMMER1.7 tumours. h, i, Quantification of intracellular IFN-γ staining by percentage in tumour (h) or spleen (i) for each indicated intratumoral lymphocyte population from mice engrafted with MC38 cells. j, k, Quantification of intracellular granzyme B staining by MFI (j) and CD107 staining in intratumoral CD8+ T cells and NK cells (k) from MC38 tumours in the indicated treatment groups. l, m, Absolute number (l) and frequency (m) of polyfunctional NK cells measured by co-staining of intracellular IFN-γ and granzyme B following ex vivo stimulation with PMA and ionomycin. Data are representative of three independent experiments with five mice per group. P values calculated using two-tailed unpaired Student’s t-tests (a), one-way ANOVA (b, f, g, j, k) or two-way ANOVA (c, e, h, i, l) and data presented as mean ± s.e.m.
Extended Data Fig. 8 DR-18 promotes intratumoral TCF1+ precursor CD8+ T cells.
Related to Fig. 3. a, Frequency of surface PD-1 and TIM3 staining on MC38 intratumoral CD44+CD8+ T cells in indicated treatment groups. b, Absolute cell number of DN (PD-1−TIM3−), SP (PD-1+TIM3−) or DP (PD-1+TIM3+) CD8+ T cells from YUMMER1.7 tumours in each treatment group. c, Frequency of intracellular TCF1 in intratumoral CD44+CD8+ T cells in MC38 tumours in each treatment group. d, Absolute number of TCF1+CD44+CD8+ T cells from MC38 tumours in each treatment group. e, Frequency of intracellular TCF1 staining in MC38 intratumoral CD44+CD8+ T cells subsets based on PD-1 and TIM3 surface expression. f, Representative histograms of intracellular TCF1 staining in intratumoral SP PD1+TIM3−CD44+CD8+ T cells from YUMMER1.7 tumours. g, Absolute number of TCF1+ SP PD1+TIM3− CD44+CD8+ T cells from YUMMER1.7 tumours in each treatment group. h, i, Frequency of intracellular IFN-γ in DP (PD-1+TIM3+) CD44+CD8+ T cells from MC38 (h) and YUMMER1.7 (i) tumours in each treatment group. j, Absolute number of TCF1+ cells in SP (PD-1+TIM3−) CD44+CD8+ T cells in draining lymph nodes from MC38 tumours in each treatment group. Data are representative of three independent experiments with five mice per group. P values were calculated using one-way (c, d, g–j) or two-way ANOVA (a, b, e) and data are presented as mean ± s.e.m.
Extended Data Fig. 9 DR-18 is effective towards MHC class I-deficient tumours.
Related to Fig. 4. a, Representative spider plots showing tumour growth in wild-type mice engrafted s.c. with MC38-B2m−/− tumours and treated with PBS, or anti-PD-1 + anti-CTLA-4, IL-18 or DR-18. b, c, Kaplan–Meier survival curves (b) and representative spider plots (c) of wild-type mice engrafted s.c. with YUMMER1.7-B2m−/− tumours and treated twice weekly with PBS, anti-PD-1 + anti-CTLA-4, IL-18, DR-18, or DR-18 + anti-CD8 and anti-NK1.1 antibodies. Depleting antibodies were given on the same schedule as for MC38-B2m−/− (see Methods). d, e, Kaplan–Meier survival curves (d) and spider plots showing tumour growth (e) of wild-type mice engrafted s.c. with 1 × 106 RMA-S tumour cells and treated with PBS, anti-PD-1, IL-18, DR-18 or DR-18 + anti-NK1.1. Spider plot data are representative of two independent experiments and survival curve data are pooled from two independent experiments. f, Absolute number of intratumoral NK cells (NK1.1+CD3−EOMES+DX5+CD200R−), NKT cells (NK1.1+CD3+) and ILC1 cells (NK1.1+CD3−EOMES−DX5−CD200R+) in wild-type mice engrafted s.c. with MC38-B2m−/− tumours in indicated treatment groups. g, t-SNE projection of different clusters in YUMMER1.7-B2m−/− tumours as a function of treatment. YUMMER1.7 tumours are shown in light grey (discussed in Fig. 3, see Methods). h, Representative flow cytometry plots of IFNγ and Ki67 staining on intratumoral NK cells from YUMMER1.7-B2m−/− tumours treated with PBS or DR-18. i, j, Frequency of Ki67+IFNγ+ NK cells from YUMMER1.7-B2m−/− (i) and MC38-B2m−/− (j) tumours treated with PBS or DR-18. k, Representative mean tumour growth in wild-type mice engrafted s.c. with MC38-B2m−/− tumours and treated with PBS, DR-18, or DR-18 + anti-FasL or anti-IFN-γ. l, Representative mean tumour growth of Prf1−/− mice engrafted s.c. with MC38-B2m−/− cells and treated with PBS (n = 3) and DR-18 (n = 5) as indicated. h–l, Data are representative of two independent experiments with five mice per group. P values calculated using two-tailed unpaired Student’s t-test and data presented as mean ± s.e.m.
Extended Data Fig. 10 Ex vivo characterization of a human DR-18 variant.
a, SPR sensorgrams of human DR-18 (hDR-18) binding to human or cynomolgus macaque (M. fascicularis) IL-18Rα and IL-18BP. b, Concentration–response curves of hIL-18 or hDR-18 on HEK Blue IL-18R reporter cells. c, Titration of hIL-18BP on a fixed concentration of hIL-18 (1 ng/ml) or hDR-18 (0.1 ng/ml) on HEK Blue IL-18R reporter cells. d, e, Concentration–response curves of IFN-γ stimulation by hIL-18 or hDR-18 on human PBMCs (d) or cynomolgus macaque PBMCs (e). IFN-γ concentration measured by ELISA. Data are representative of at least two independent experiments with n = 2 for b–e and presented as mean ± s.e.m.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Zhou, T., Damsky, W., Weizman, OE. et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 583, 609–614 (2020). https://doi.org/10.1038/s41586-020-2422-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2422-6
This article is cited by
-
The 4th NextGen therapies of SJIA and MAS, part 4: it is time for IL-18 based trials in systemic juvenile idiopathic arthritis?
Pediatric Rheumatology (2024)
-
Phenome-wide Mendelian randomisation analysis of 378,142 cases reveals risk factors for eight common cancers
Nature Communications (2024)
-
Dictionary of immune responses to cytokines at single-cell resolution
Nature (2024)
-
Biological and clinical roles of IL-18 in inflammatory diseases
Nature Reviews Rheumatology (2024)
-
The role of pyroptosis and gasdermin family in tumor progression and immune microenvironment
Experimental Hematology & Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.