Abstract
Synucleinopathies, which include multiple system atrophy (MSA), Parkinson’s disease, Parkinson’s disease with dementia and dementia with Lewy bodies (DLB), are human neurodegenerative diseases1. Existing treatments are at best symptomatic. These diseases are characterized by the presence of, and believed to be caused by the formation of, filamentous inclusions of α-synuclein in brain cells2,3. However, the structures of α-synuclein filaments from the human brain are unknown. Here, using cryo-electron microscopy, we show that α-synuclein inclusions from the brains of individuals with MSA are made of two types of filament, each of which consists of two different protofilaments. In each type of filament, non-proteinaceous molecules are present at the interface of the two protofilaments. Using two-dimensional class averaging, we show that α-synuclein filaments from the brains of individuals with MSA differ from those of individuals with DLB, which suggests that distinct conformers or strains characterize specific synucleinopathies. As is the case with tau assemblies4,5,6,7,8,9, the structures of α-synuclein filaments extracted from the brains of individuals with MSA differ from those formed in vitro using recombinant proteins, which has implications for understanding the mechanisms of aggregate propagation and neurodegeneration in the human brain. These findings have diagnostic and potential therapeutic relevance, especially because of the unmet clinical need to be able to image filamentous α-synuclein inclusions in the human brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Raw cryo-EM micrographs are available in EMPIAR, entry numbers EMPIAR-10357 (MSA case 1) and EMPIAR-10358 (MSA case 2). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-10650 for type I filaments from MSA case 1, EMD-10651 for type II1 filaments from MSA case 2 and EMD-10652 for type II2 filaments from MSA case 2. The corresponding atomic models have been deposited in the Protein Data Bank under the following accession numbers: 6XYO for type I filaments from MSA case 1, 6XYP for type II1 filaments from MSA case 2 and 6XYQ for type II2 filaments from MSA case 2. LC–MS/MS data were obtained from the ProteomeXchange database and have been deposited in Japan Proteome Standard Repository/Database (JPOST) under the identifier PXD018434.
References
Goedert, M., Jakes, R. & Spillantini, M. G. The synucleinopathies: twenty years on. J. Parkinsons Dis. 7, S51–S69 (2017).
Spillantini, M. G. et al. α-Synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Polymeropoulos, M. H. et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017).
Falcon, B. et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140 (2018).
Falcon, B. et al. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 136, 699–708 (2018).
Falcon, B. et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019).
Zhang, W. et al. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 8, e43584 (2019).
Zhang, W. et al. Novel tau filament fold in corticobasal degeneration. Nature 580, 283–287 (2020).
Singleton, A. B. et al. α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003).
Kiely, A. P. et al. α-Synucleinopathy associated with G51D SNCA mutation: a link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol. 125, 753–769 (2013).
Kiely, A. P. et al. Distinct clinical and neuropathological features of G51D SNCA mutation cases compared with SNCA duplication and H50Q mutation. Mol. Neurodegener. 10, 41 (2015).
Pasanen, P. et al. Novel α-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol. Aging 35, 2180.e1–2180. e5 (2014).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014).
Dejerine, J. & Thomas, A. L’atrophie olivo-ponto-cérébelleuse. Nouv. Iconogr. Salpêtrière 13, 330–370 (1900).
Graham, J. G. & Oppenheimer, D. R. Orthostatic hypotension and nicotine sensitivity in a case of multiple system atrophy. J. Neurol. Neurosurg. Psychiatry 32, 28–34 (1969).
Quinn, N. Multiple system atrophy – the nature of the beast. J. Neurol. Neurosurg. Psychiatry 52 (suppl.), 78–89 (1989).
Papp, M. I., Kahn, J. E. & Lantos, P. L. Glial cytoplasmic inclusions in the CNS of patients with multiple system atrophy (striatonigral degeneration, olivopontocerebellar atrophy and Shy–Drager syndrome). J. Neurol. Sci. 94, 79–100 (1989).
Wakabayashi, K., Yoshimoto, M., Tsuji, S. & Takahashi, H. α-synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci. Lett. 249, 180–182 (1998).
Spillantini, M. G. et al. Filamentous α-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci. Lett. 251, 205–208 (1998).
Tu, P. H. et al. Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble α-synuclein. Ann. Neurol. 44, 415–422 (1998).
Kato, S. & Nakamura, H. Cytoplasmic argyrophilic inclusions in neurons of pontine nuclei in patients with olivopontocerebellar atrophy: immunohistochemical and ultrastructural studies. Acta Neuropathol. 79, 584–594 (1990).
Petrovic, I. N. et al. Multiple system atrophy–parkinsonism with slow progression and prolonged survival: a diagnostic catch. Mov. Disord. 27, 1186–1190 (2012).
Davidson, W. S., Jonas, A., Clayton, D. F. & George, J. M. Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273, 9443–9449 (1998).
Uéda, K. et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl Acad. Sci. USA 90, 11282–11286 (1993).
Li, H. T., Du, H. N., Tang, L., Hu, J. & Hu, H. Y. Structural transformation and aggregation of human α-synuclein in trifluoroethanol: non-amyloid component sequence is essential and β-sheet formation is prerequisite to aggregation. Biopolymers 64, 221–226 (2002).
Crowther, R. A., Jakes, R., Spillantini, M. G. & Goedert, M. Synthetic filaments assembled from C-terminally truncated α-synuclein. FEBS Lett. 436, 309–312 (1998).
Conway, K. A., Harper, J. D. & Lansbury, P. T. Jr. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry 39, 2552–2563 (2000).
Serpell, L. C., Berriman, J., Jakes, R., Goedert, M. & Crowther, R. A. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc. Natl Acad. Sci. USA 97, 4897–4902 (2000).
Miake, H., Mizusawa, H., Iwatsubo, T. & Hasegawa, M. Biochemical characterization of the core structure of α-synuclein filaments. J. Biol. Chem. 277, 19213–19219 (2002).
Mougenot, A. L. et al. Prion-like acceleration of a synucleinopathy in a transgenic mouse model. Neurobiol. Aging 33, 2225–2228 (2012).
Luk, K. C. et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012).
Masuda-Suzukake, M. et al. Prion-like spreading of pathological α-synuclein in brain. Brain 136, 1128–1138 (2013).
Osterberg, V. R. et al. Progressive aggregation of α-synuclein and selective degeneration of Lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Rep. 10, 1252–1260 (2015).
Peelaerts, W. et al. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015).
Peng, C. et al. Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018).
Prusiner, S. B. et al. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl Acad. Sci. USA 112, E5308–E5317 (2015).
Tarutani, A., Arai, T., Murayama, S., Hisanaga, S. I. & Hasegawa, M. Potent prion-like behaviors of pathogenic α-synuclein and evaluation of inactivation methods. Acta Neuropathol. Commun. 6, 29 (2018).
Yamasaki, T. R. et al. Parkinson’s disease and multiple system atrophy have distinct α-synuclein seed characteristics. J. Biol. Chem. 294, 1045–1058 (2019).
Lavenir, I. et al. Silver staining (Campbell–Switzer) of neuronal α-synuclein assemblies induced by multiple system atrophy and Parkinson’s disease brain extracts in transgenic mice. Acta Neuropathol. Commun. 7, 148 (2019).
Klingstedt, T. et al. Luminescent conjugated oligothiophenes distinguish between α-synuclein assemblies of Parkinson’s disease and multiple system atrophy. Acta Neuropathol. Commun. 7, 193 (2019).
Strohäker, T. et al. Structural heterogeneity of α-synuclein fibrils amplified from patient brain extracts. Nat. Commun. 10, 5535 (2019).
Shahnawaz, M. et al. Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277 (2020).
Campbell, B. C. V. et al. The solubility of α-synuclein in multiple system atrophy differs from that of dementia with Lewy bodies and Parkinson’s disease. J. Neurochem. 76, 87–96 (2001).
Fujiwara, H. et al. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 (2002).
Spillantini, M. G., Crowther, R. A., Jakes, R., Hasegawa, M. & Goedert, M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl Acad. Sci. USA 95, 6469–6473 (1998).
Tuttle, M. D. et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016).
Guerrero-Ferreira, R. et al. Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018).
Li, Y. et al. Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018).
Li, B. et al. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018).
Guerrero-Ferreira, R. et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019).
Boyer, D. R. et al. Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 26, 1044–1052 (2019).
Boyer, D. R. et al. The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl Acad. Sci. USA 117, 3592–3602 (2020).
Zhao, K. et al. Parkinson’s disease associated mutation E46K of α-synuclein triggers the formation of a novel fibril structure. Preprint at https://www.biorxiv.org/content/10.1101/870758v2.
Sangwan, S. et al. Inhibition of synucleinopathic seeding by rationally designed inhibitors. eLife 9, e46775 (2020).
Goedert, M., Falcon, B., Zhang, W., Ghetti, B. & Scheres, S. H. W. Distinct conformers of assembled tau in Alzheimer’s and Pick’s diseases. Cold Spring Harb. Symp. Quant. Biol. 83, 163–171 (2018).
Goedert, M., Spillantini, M. G., Cairns, N. J. & Crowther, R. A. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 (1992).
Giasson, B. I. et al. A panel of epitope-specific antibodies detects protein domains distributed throughout human α-synuclein in Lewy bodies of Parkinson’s disease. J. Neurosci. Res. 59, 528–533 (2000).
Spina, S. et al. The tauopathy associated with mutation +3 in intron 10 of Tau: characterization of the MSTD family. Brain 131, 72–89 (2008).
Nonaka, T., Watanabe, S. T., Iwatsubo, T. & Hasegawa, M. Seeded aggregation and toxicity of α-synuclein and tau: cellular models of neurodegenerative diseases. J. Biol. Chem. 285, 34885–34898 (2010).
Tarutani, A. et al. The effect of fragmented pathogenic α-synuclein seeds on prion-like propagation. J. Biol. Chem. 291, 18675–18688 (2016).
Kametani, F. et al. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017).
Scheres, S. H. W. Amyloid structure determination in RELION-3.1. Acta Crystallogr. D. 76, 94–101 (2020).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Acknowledgements
We thank the families of the patients for donating brain tissues; T. Nakane for help with RELION; T. Darling and J. Grimmett for help with high-performance computing; F. Epperson, U. Kuederli, R. Otani and R. M. Richardson for support with neuropathology; R. A. Crowther, B. Falcon, S. Lovestam, M. G. Spillantini and W. Zhang for helpful discussions. M.G. is an Honorary Professor in the Department of Clinical Neurosciences of the University of Cambridge and an Associate Member of the UK Dementia Research Institute. This work was supported by the UK Medical Research Council (MC_UP_A025_1013, to S.H.W.S., and MC_U105184291, to M.G.), Eli Lilly and Company (to M.G.), the European Union (EU/EFPIA/Innovative Medicines Initiative [2] Joint Undertaking IMPRIND, project 116060, to M.G.), the Japan Agency for Medical Research and Development (JP18ek0109391 and JP18dm020719, to M.H.), the US National Institutes of Health (P30-AG010133 and U01-NS110437, to B.G.) and the Department of Pathology and Laboratory Medicine, Indiana University School of Medicine (to B.G.). We acknowledge Y. Chaban at Diamond for access to and support from the cryo-EM facilities at the UK electron Bio-Imaging Centre (eBIC), proposal EM17434-75, funded by the Wellcome Trust, the MRC and the BBSRC, for acquisition of the MSA case 1 dataset. This study was supported by the MRC-LMB electron microscopy facility.
Author information
Authors and Affiliations
Contributions
A.T., B.G., T.M., T.T., T.A., K.H., S.M., M.Y. and M.H. identified patients, performed neuropathology and extracted α-synuclein filaments from MSA cases 1–5 and DLB cases 1 and 2; M.S. extracted α-synuclein filaments from DLB case 3 and conducted immunolabelling of filaments from MSA cases 1–5 and DLB cases 1–3; F.K. and M.H. carried out mass spectrometry; A.T. and M.H. did seeded aggregation; M.S. and Y.S. performed cryo-EM; Y.S., M.S. and S.H.W.S. analysed the cryo-EM data; Y.S. and A.G.M. built the atomic models; S.H.W.S. and M.G. supervised the project; all authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks David Eliezer, Henning Stahlberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Filamentous α-synuclein pathology and immunolabelling of α-synuclein filaments from MSA.
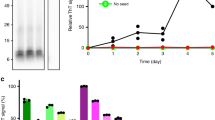
a, Staining of inclusions in the frontal cortex in MSA cases 1, 2, 3 and 5 and the cerebellum in MSA case 1 by the antibody pS129 (brown). Scale bar, 50 μm. b, Negative-stain electron microscopy images of filaments from the frontal cortex in MSA cases 1, 2, 3 and 5, and the cerebellum in MSA case 1. Scale bar, 50 nm. c, d, Representative immunogold negative-stain electron microscopy images of α-synuclein filaments extracted from the frontal cortex in MSA cases 1, 2, 3 and 5, the cerebellum in MSA case 1 and the putamen in MSA cases 1–5. Filaments were labelled with the antibody PER4. Scale bar, 200 nm. e, Immunoblots of sarkosyl-insoluble material from the putamen for MSA cases 1–5, using the anti-α-synuclein antibodies Syn303 (N terminus), PER4 (C terminus) and pS129 (phosphorylation of S129). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Aggregation of α-synuclein in SH-SY5Y cells after addition of seeds from the putamen for MSA cases 1–5.
Quantification of wild-type human α-synuclein phosphorylated at S129 in SH-SY5Y cells after addition of variable amounts of α-synuclein seeds from the putamen for MSA cases 1–5. The results are expressed as mean ± s.e.m. (n = 3 experiments).
Extended Data Fig. 3 Cryo-EM images and 2D classification of MSA filaments.
a, c, Representative cryo-EM images of α-synuclein filaments from the putamen in MSA cases 1–5, the frontal cortex in MSA cases 1, 2, 3 and 5 and the cerebellum in MSA case 1. Scale bar, 100 nm. b, d, Two-dimensional class averages spanning an entire crossover of type I and type II filaments extracted from the putamen in MSA cases 1–5, the frontal cortex in MSA cases 1, 2, 3 and 5, and the cerebellum in MSA case 1.
Extended Data Fig. 4 Evaluation of the resolution of cryo-EM maps and of refined models.
a–c, For type I (a), type II1 (b) and type II2 (c) filaments of α-synuclein from MSA: Fourier shell correlation (FSC) curves of two independently refined half-maps (black line); FSC curves of final cryo-EM reconstruction and refined atomic model (red); FSC curves of first half-map and the atomic model refined against this map (blue); FSC curves of second half-map and the atomic model refined against the first half-map (yellow dashes). d–f, Estimates of local resolution of the reconstructions of type I (d), type II1 (e) and type II2 (f) filaments of α-synuclein from MSA.
Extended Data Fig. 5 Type I and type II filaments of α-synuclein from MSA.
a, Schematic of a type I filament, showing asymmetric PF-IA and PF-IB. The non-proteinaceous density at the protofilament interface is shown in light red. b, Schematic of a type II filament, showing asymmetric PF-IIA and PF-IIB. The non-proteinaceous density at the protofilament interface is shown in light red.
Extended Data Fig. 6 The inter-protofilament interfaces of MSA type I and type II α-synuclein filaments.
a, b, Rendered view of secondary structure elements in MSA type I (a) and type II (b) protofilament folds perpendicular to the helical axis of inter-protofilament interfaces, depicted as three rungs. Because of variations in the height of both polypeptide chains along the helical axis, each α-synuclein molecule interacts with three different molecules in the opposing protofilament. If one considers the interaction between two opposing molecules to be on the same β-sheet rung in the central cavity, the N-terminal arm of PF-IA or PF-IIA interacts with the C-terminal body of the PF-IB or PF-IIB molecule, which is one rung higher, while the C-terminal body of PF-IA or PF-IIA interacts with the N-terminal arm of the PF-IB or PF-IIB molecule, which is one rung lower. c, d, Compatibility of mutant α-synuclein (G51D and A53E) with MSA type I and type II filaments. Close-up views of atomic models of type I (c) and type II (d) α-synuclein folds containing D51 (cyan) and E53 (green). Each mutation adds two negatively charged side chains per rung in the second shell of residues around the central cavity, thus reducing the net positive charge of the shell.
Extended Data Fig. 7 Filamentous α-synuclein pathology in DLB.
a, Staining of inclusions in the frontal cortex in DLB cases 1 and 2 and the amygdala in DLB case 3 by the antibody pS129 (brown). Scale bar, 50 μm. b, Negative-stain electron microscopy images of filaments from the frontal cortex in DLB cases 1 and 2 and the amygdala in DLB case 3. Scale bar, 50 nm. c, Representative immunogold negative-stain electron microscopy images of α-synuclein filaments extracted from the frontal cortex in DLB cases 1 and 2 and the amygdala in DLB case 3. Filaments were labelled with the antibody PER4, which recognizes the C terminus of α-synuclein. Arrowheads point to an unlabelled tau paired helical filament. Scale bar, 200 nm. d, Representative cryo-EM images of α-synuclein filaments from the frontal cortex in DLB cases 1 and 2, and the amygdala in DLB case 3. Scale bar, 200 nm. Arrowheads point to a tau paired helical filament, as evidenced by a three-dimensional reconstruction (inset), calculated as previously described6. e, Two-dimensional class averages of α-synuclein filaments extracted from the frontal cortex in DLB cases 1 and 2 and the amygdala in DLB case 3.
Extended Data Fig. 8 Structures of α-synuclein protofilament cores.
a, Schematic of secondary structure elements in the α-synuclein protofilament cores of MSA. Red arrows point to the non-proteinaceous density (in light red) at protofilament interfaces. b, c, Secondary structure elements in the α-synuclein protofilament cores assembled from recombinant wild-type (b) and mutant (c) α-synuclein. β-Strands are shown as thick arrows. d, Schematic depicting the first 100 amino acids of human α-synuclein, comparing secondary structure elements in protofilament cores from MSA with those in protofilament cores assembled from recombinant α-synuclein. As observed previously for tau filaments9, the arrangement of residues in β-strands is largely conserved among protofilament cores. This is especially the case for residues that adopt the conserved three-layered L-shaped motif, and less so for residues in the N-terminal arms.
Extended Data Fig. 9 MSA filaments differ from those assembled with recombinant α-synuclein.
a, Overlay of the three-layered L-shaped motifs of MSA α-synuclein filaments (yellow, orange, pink and purple) and filaments assembled in vitro using recombinant α-synuclein that contain a similar motif (grey). Despite topological similarities, none of the three-layered L-shaped motifs in recombinant α-synuclein protofilaments is identical to those of MSA protofilaments. The closest similarity to an in vitro structure is between PF-IIB2 and PDB 6PEO52, which differ only in the bend positions in the outer layer (between E57 and K58 for PF-IIB2 and between T59 and K60 for PDB 6PEO). b, Overlay of MSA and recombinant α-synuclein structures on the basis of the turn at residues K43–V52, revealing a conserved interface between residues E46–V49 and V74–A78 or A76–K80 (red highlight), including the formation of a salt bridge between E46 and K80. c, Overlay of MSA and recombinant α-synuclein structures on the basis of the conserved turn at residues V63–T72, revealing a second conserved turn (V63–T72) and a conserved packing through tight interdigitations of small side chains between residues A69–T72 and residues on the inner layer (green highlight). In MSA PF-IA and PF-IIA filaments, as well as in PDB 6OSM47, these residues are A89 and A91; in MSA PF-IB and PF-IIB filaments, as well as in PDB 6PEO, they are G93 and V95; in several recombinant α-synuclein structures, they are A91 and G93.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3 and Supplementary Figure 1.
Peer Review File
Reviewer reports and authors' response from the peer review of this Article at Nature Communications.
Rights and permissions
About this article
Cite this article
Schweighauser, M., Shi, Y., Tarutani, A. et al. Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). https://doi.org/10.1038/s41586-020-2317-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2317-6
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Phosphorylation and O-GlcNAcylation at the same α-synuclein site generate distinct fibril structures
Nature Communications (2024)
-
RNA as a component of scrapie fibrils
Scientific Reports (2024)
-
Structural polymorphism of amyloid fibrils in ATTR amyloidosis revealed by cryo-electron microscopy
Nature Communications (2024)
-
O-GlcNAc forces an α-synuclein amyloid strain with notably diminished seeding and pathology
Nature Chemical Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.