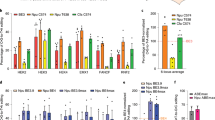
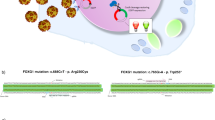
Abstract
Hutchinson–Gilford progeria syndrome (HGPS or progeria) is typically caused by a dominant-negative C•G-to-T•A mutation (c.1824 C>T; p.G608G) in LMNA, the gene that encodes nuclear lamin A. This mutation causes RNA mis-splicing that produces progerin, a toxic protein that induces rapid ageing and shortens the lifespan of children with progeria to approximately 14 years1,2,3,4. Adenine base editors (ABEs) convert targeted A•T base pairs to G•C base pairs with minimal by-products and without requiring double-strand DNA breaks or donor DNA templates5,6. Here we describe the use of an ABE to directly correct the pathogenic HGPS mutation in cultured fibroblasts derived from children with progeria and in a mouse model of HGPS. Lentiviral delivery of the ABE to fibroblasts from children with HGPS resulted in 87–91% correction of the pathogenic allele, mitigation of RNA mis-splicing, reduced levels of progerin and correction of nuclear abnormalities. Unbiased off-target DNA and RNA editing analysis did not detect off-target editing in treated patient-derived fibroblasts. In transgenic mice that are homozygous for the human LMNA c.1824 C>T allele, a single retro-orbital injection of adeno-associated virus 9 (AAV9) encoding the ABE resulted in substantial, durable correction of the pathogenic mutation (around 20–60% across various organs six months after injection), restoration of normal RNA splicing and reduction of progerin protein levels. In vivo base editing rescued the vascular pathology of the mice, preserving vascular smooth muscle cell counts and preventing adventitial fibrosis. A single injection of ABE-expressing AAV9 at postnatal day 14 improved vitality and greatly extended the median lifespan of the mice from 215 to 510 days. These findings demonstrate the potential of in vivo base editing as a possible treatment for HGPS and other genetic diseases by directly correcting their root cause.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
Code used in this study is available from https://github.com/CwilsonBroad/Koblan_2020_In-Vivo-Adenine-Base-Editing-Corrects-Hutchinson-Gilford-Progeria-Syndrome (Supplementary Note 3).
References
Eriksson, M. et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423, 293–298 (2003).
De Sandre-Giovannoli, A. et al. Lamin A truncation in Hutchinson–Gilford progeria. Science 300, 2055 (2003).
Gordon, L. B. et al. Impact of farnesylation inhibitors on survival in Hutchinson–Gilford progeria syndrome. Circulation 130, 27–34 (2014).
Gordon, L. B., Brown, W. T. & Collins, F. S. Hutchinson–Gilford Progeria Syndrome (GeneReviews, 2019).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Olive, M. et al. Cardiovascular pathology in Hutchinson–Gilford progeria: correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 30, 2301–2309 (2010).
Gerhard-Herman, M. et al. Mechanisms of premature vascular aging in children with Hutchinson–Gilford progeria syndrome. Hypertension 59, 92–97 (2012).
Rivera-Torres, J. et al. Cardiac electrical defects in progeroid mice and Hutchinson–Gilford progeria syndrome patients with nuclear lamina alterations. Proc. Natl Acad. Sci. USA 113, E7250–E7259 (2016).
Prakash, A. et al. Cardiac abnormalities in patients with Hutchinson–Gilford progeria syndrome. JAMA Cardiol. 3, 326–334 (2018).
Stehbens, W. E., Wakefield, S. J., Gilbert-Barness, E., Olson, R. E. & Ackerman, J. Histological and ultrastructural features of atherosclerosis in progeria. Cardiovasc. Pathol. 8, 29–39 (1999).
Gordon, L. B. et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 109, 16666–16671 (2012).
Capell, B. C. & Collins, F. S. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 7, 940–952 (2006).
Lai, W.-F. & Wong, W.-T. Progress and trends in the development of therapies for Hutchinson–Gilford progeria syndrome. Aging Cell 19, e13175 (2020).
Beyret, E. et al. Single-dose CRISPR–Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat. Med. 25, 419–422 (2019).
Santiago-Fernández, O. et al. Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat. Med. 25, 423–426 (2019).
Suzuki, K. et al. Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction. Cell Res. 29, 804–819 (2019).
Scaffidi, P. & Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nat. Med. 11, 440–445 (2005).
Jiang, F. & Doudna, J. A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529 (2017).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Varga, R. et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 103, 3250–3255 (2006).
Huang, T. P. et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631 (2019).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Eisenberg, E. & Levanon, E. Y. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018).
Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97–110 (2020).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Inagaki, K. et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14, 45–53 (2006).
Bostick, B., Ghosh, A., Yue, Y., Long, C. & Duan, D. Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Ther. 14, 1605–1609 (2007).
Wang, L., Wang, H., Bell, P., McMenamin, D. & Wilson, J. M. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum. Gene Ther. 23, 533–539 (2012).
Kessler, P. D. et al. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc. Natl Acad. Sci. USA 93, 14082–14087 (1996).
Nathwani, A. C. et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014).
Hagan, C. When are mice considered old? The Jackson Laboratory https://www.jax.org/news-and-insights/jax-blog/2017/november/when-are-mice-considered-old (2017).
Chandler, R. J., Sands, M. S. & Venditti, C. P. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum. Gene Ther. 28, 314–322 (2017).
Nault, J.-C. et al. Wild-type AAV insertions in hepatocellular carcinoma do not inform debate over genotoxicity risk of vectorized AAV. Mol. Ther. 24, 660–661 (2016).
Geng, D., Zhao, W., Feng, Y. & Liu, J. Overexpression of Rab25 promotes hepatocellular carcinoma cell proliferation and invasion. Tumour Biol. 37, 7713–7718 (2016).
Donsante, A. et al. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther. 8, 1343–1346 (2001).
Donsante, A. et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science 317, 477 (2007).
Embury, J. E., Charron, C. C., Poirier, A. E. & Zori, A. Long term portal vein administration of AAV-WPRE vector results in increased incidence of neoplastic disease and hepatic pathology. Mol. Ther. 13, S83 (2006).
Sands, M. S. AAV-mediated liver-directed gene therapy. Methods Mol. Biol. 807, 141–157 (2011).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with enhanced Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Osorio, F. G. et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 3, 106ra107 (2011).
Mateos, J. et al. Next-generation sequencing and quantitative proteomics of Hutchinson–Gilford progeria syndrome-derived cells point to a role of nucleotide metabolism in premature aging. PloS One 13, e0205878 (2018).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Wu, D. & Smyth, G. K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012).
Cao, K. et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson–Gilford progeria syndrome cells. Sci. Transl. Med. 3, 89ra58 (2011).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Frengen, E. et al. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58, 250–253 (1999).
DuBose, A. J. et al. Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene. Nucleic Acids Res. 41, e70
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Acknowledgements
This work was supported by NIH grants U01AI142756, UG3AI150551, UG3TR002636, RM1HG009490, R01EB022376, R35GM118062, Z01HG200305, R01HL146654 and R01HL126784, and by the Howard Hughes Medical Institute. L.W.K. acknowledges an NSF GRFP fellowship; C.W. acknowledges a Damon Runyon Cancer Research Foundation fellowship (DRG-2343-18); G.A.N. acknowledges a Helen Hay Whitney postdoctoral fellowship; and L.B.G., L.W.K. and D.R.L. acknowledge support from The Progeria Research Foundation.
Author information
Authors and Affiliations
Contributions
L.W.K., J.D.B., C.Y.L., M.R.E., F.S.C. and D.R.L. designed the research. L.W.K., M.R.E., C.W., W.A.C., L.D., Y.G.G., X.M., G.A.N., S.P.D., J.D.B. performed cell culture experiments. L.W.K., M.R.E., C.W., W.A.C., J.M.L., Z.X., U.L.T., L.D., S.P.D., N.N., Q.S., C.K. and J.D.B. performed in vivo experiments. L.B.G., K.C., F.S.C., J.D.B. and D.R.L. supervised the project. L.W.K. and D.R.L. wrote the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
D.R.L. is a co-founder of Beam Therapeutics, Prime Medicine, Pairwise Plants and Editas Medicine, companies that use genome editing. The remaining authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Additional characterization of patient-derived cells treated with ABE7.10max-VRQR.
a, Bystander V690A editing in patient-derived HGADFN167 and HGADFN188 cells 20 days after treatment with lentiviral ABE7.10max-VRQR. b, Indel formation frequency at the c.1824 target locus in HGADFN167 and HGADFN188 cells 20 days after treatment with lentiviral ABE7.10max-VRQR. Data are mean ± s.d. for n = 5 technical replicates (individual points) for a, b. c, Sanger DNA sequencing traces of untreated HGADFN167 cells, 20-day treated HGADFN167 cells and unaffected control cells. The target nucleotide is boxed. d, qPCR-normalized progerin mRNA abundance in cells described in c. Data are mean ± s.d. for n = 3 biological replicates. e, Western blot analysis of HGADFN167 cells described in c. LMNA, progerin, and LMNC protein are all stained on the gel, A GAPDH loading control is shown below. An additional replicate is provided in Fig. 1d. Unaff. ctrl, control cells from an unaffected parent. f, Sanger DNA sequencing traces of untreated, non-targeting (NT)-sgRNA-treated and ABE-treated HGADFN155 fibroblasts at a 20-day time point to ensure NT-sgRNA did not lead to DNA editing. g, Western blot analysis of cells described in f as well as treated cells at a 30-day time point. LMNA, progerin, and LMNC proteins are all stained on the gel, a β-actin loading control is shown below. Expected molecular weights: lamin A, 74 kDa; progerin, 69 kDa; lamin C, 65 kDa. Complete blots are available in Supplementary Fig. 1. Additional replication was not performed. h, HGADFN167 (left) and HGADFN188 (right) cell lines untreated or treated with lentiviral ABE7.10max-VRQR after 10 or 20 days show similar relative distributions of A-to-I SNVs in their transcriptomes compared with the hg38 human genome reference sequence. On average, 36 ± 3.6% of SNPs in these samples occur with around 100% frequency, suggesting they arise from genomic sequence variations; however, we cannot explicitly exclude them from consideration as no whole-genome sequence is available for these cell lines. Raw counts of 100% edited SNPs per sample are: untreated HGADFN167 cells (849), HGADFN167 10 d after treatment (883), HGADFN167 20 d after treatment (871), untreated HGADFN188 cells (488), HGADFN188 10 d after treatment (501), HGADFN188 20 d after treatment (510).
Extended Data Fig. 2 CIRCLE-seq analysis of HGADFN167 and HGADFN188 cells using Cas9-VRQR and the progeria-targeting sgRNA.
a, b, CIRCLE-seq read counts for Cas9-VRQR nuclease-treated genomic DNA from HGADFN167 (a) and HGADFN188 (b) cell lines. Targeted amplicon sequencing was used to assess the off-target base editing for 36 noted total loci distributed across both cell lines. DNA at 32 of 35 loci amplified efficiently from both cell lines (denoted by black check marks), DNA at 3 loci failed to amplify (denoted by red X marks). Complete CIRCLE-seq data are provided in Supplementary Data 4. c, SDS–PAGE gel stained with InstantBlue to follow protein purification of Cas9-VRQR. Totals of 0.5 μl clarified lysate, 0.25 μl nickel column elution or 0.1 μl of the concentrated protein stock after His-tag purification and ion-exchange chromatography were added to 5 μl NuPAGE loading buffer. Samples were denatured at 98 °C for 5 min before loading onto the 4–12% acrylamide gel. A Precision Plus Protein Kaleidoscope Pre-Stained Ladder (Bio-Rad) was used as reference. The desired Cas9-VRQR has a predicted molecular weight of 161.9 kDa. Additional replication was not performed.
Extended Data Fig. 3 DNA on-target editing, bystander editing and indel efficiencies across tissues from in vivo injection route optimization experiments.
a, Dual AAV9 encoding split-intein ABE7.10max-VRQR base editor halves and the LMNA-targeting sgRNA were injected into homozygous LMNA c.1824 C>T mice. P3 retro-orbital (RO) injections (5 × 1010 vg of each AAV, 1 × 1011 vg total), P14 RO injections (5 × 1011 vg of each AAV, 1 × 1012 vg total) and P14 intraperitoneal (IP) injections (5 × 1011 vg of each AAV, 1 × 1012 vg total) were tested. At 6-weeks of age, mice were euthanized and heart, muscle, liver, aorta and bone were isolated for sequencing analysis. Tissues were sub-sectioned for sequencing analysis to ensure sub-sections did not show differences in editing efficiencies for downstream analyses. Each bar represents a different tissue subsection. DNA-editing efficiencies correcting LMNA c.1824 from T (pathogenic) to C (wild-type) for P3 RO-injected mice (left, n = 4), P14 RO-injected mice (middle, n = 5) and P14 IP-injected mice (right, n = 5) at 6 weeks of age are shown for five disease-relevant tissues. Data are mean ± s.d. b, Apparent LMNA c.1824 T (pathogenic) to C (wild-type) mutations from tissue samples of saline-injected P3 RO (left) and P14 RO (right) control mice at 6 months of age show background signal due to amplicon crossover during PCR between the human diseased allele and the wild-type mouse allele, which share 90% overall sequence identity within the amplified region. Similar crossover levels were observed across 11 tissues in both P3 RO and P14 RO saline-injected mice. Data are mean ± s.d. for n = 12 mice (6 male, 6 female). c, Computational filtering of same sequencing reads shown in b after removing any reads containing any mouse-specific sequence variations, analysing only reads containing exclusively human sequence. The script used to remove mouse-containing sequencing reads is in the link in Supplementary Note 3 and is described in the Methods (‘High-throughput sequencing of in vivo samples’). d, DNA editing for P3- and P14-injected mice at 6 months of age across 11 tissues. Each point represents a biological replicate of a tissue taken from a unique mouse (n = 12 for each group). e, V690A bystander editing frequency across eleven tissues for P3 RO and P14 RO ABE-treated mice at 6 months of age (n = 12 for each group). f, Indel frequencies at the c.1824 target locus across 11 tissues for P3 RO and P14 RO ABE-treated mice at 6 months of age. Data are mean ± s.d. for the indicated number of biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Student’s unpaired two-sided t-test.
Extended Data Fig. 4 Quantification of LMNA and progerin transcript abundance by ddPCR in mice that were retro-orbitally injected with saline or ABE-AAV9 at P3 or P14.
a, ddPCR counts for LMNA (grey bars) and progerin (red bars) RNA transcript abundance in P3 RO saline- and ABE-AAV9-injected mice. Data are mean ± s.d. for n = 12 mice. b, ddPCR counts for LMNA (grey bars) and progerin (red bars) RNA transcript abundance in P14 RO saline- and ABE-AAV9-injected mice. Data are mean ± s.d. for n = 12 biological replicates for all samples except for saline-injected mouse skin (n = 11), WAT (n = 7), visceral fat (n = 11), tibia (n = 11) and aorta (n = 8); and ABE-AAV9-injected mouse WAT (n = 11), tibia (n = 9) and aorta (n = 10). Visc. fat, visceral fat. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Student’s unpaired two-sided t-test for a, b. Liver and heart values are reproduced from Fig. 3c for ease of comparison.
Extended Data Fig. 5 Quantification of western blots.
Liver, heart and aorta tissue western blots for P3-injected (top half of each tissue set) and P14-injected mice (bottom half of each tissue set) were quantified by western blot. Samples from females appear in the left column, and samples from males are in the right column. Each lane represents the tissue type specified on the left taken from a different mouse. Control mice were treated with saline instead of ABE-AAV9. WT indicates C57BL/6 mouse lacking the transgene, showing that the antibody is specific to human lamin proteins and progerin. The abundance of lamin A or progerin protein relative to β-actin in saline- or ABE-treated mouse tissues was quantified by normalizing the fluorescence signal from the secondary antibody for each band (800 nm for progerin and lamins, and 680 nm for actin; see Methods (‘Protein isolation and western blotting from mouse tissues’). The normalized protein abundance relative to saline-treated samples (set to 100) is shown in the bar graphs. Control mice were treated identically to the corresponding ABE-treated mice except injected with saline instead of ABE-AAV9. Raw fluorescent signal for progerin protein measured at the 800-nm wavelength (using IRDye-labelled antibody) is displayed under each lane. Data are mean ± s.d. for n = 5 or 6 biological replicates, as indicated. The n for each sample type is listed in each figure panel. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Student’s unpaired two-sided t-test. Expected molecular weights: lamin A, 74 kDa; progerin, 69 kDa; lamin C, 65 kDa. Liver and heart blots are reproduced from Fig. 3c for ease of comparison. Complete blots are available in Supplementary Fig. 1.
Extended Data Fig. 6 Aortic histology of male and female mice that were retro-orbitally injected with saline or ABE-AAV9 at P3.
a, Representative aorta cross-sections for P3 RO saline- or ABE-AAV9-injected males at 6 months of age. Left images were stained with H&E; right images were stained with Movat’s pentachrome stain. b, Representative aorta cross-sections for P3 RO saline- or ABE-AAV9-treated females at 6 months of age. Left images were stained with H&E; right images were stained with Movat’s pentachrome stain. Unaffected WT are wild-type C57BL/6 mice. WT M2, 9609M, 9177M, WT F3, 9628F and 9148F are reproduced from Fig. 4 for ease of comparison. These sections each represent replicates from different mice.
Extended Data Fig. 7 Aortic histology of male and female mice that were retro-orbitally injected with saline or ABE-AAV9 at P14.
a, Representative aorta cross-sections for P14 RO saline- or ABE-AAV9-injected males at 6 months of age. Left images were stained with H&E; right images were stained with Movat’s pentachrome stain. b, Representative aorta cross-sections for P14 RO saline- or ABE-AAV9-treated females at 6 months of age. Left images were stained with H&E; right images were stained with Movat’s pentachrome stain. Unaffected WT are wild-type C57BL/6 mice, reproduced from Extended Data Fig. 9a, b for ease of comparison. Images from the following mice are reproduced from Fig. 4 for ease of comparison: WT M2, 9440M, 9459M, WT F3, 9536F and 9535F. These sections each represent replicates from different mice.
Extended Data Fig. 8 Immunofluorescence staining of aortas from male and female control C57BL/6 mice, untreated mice and mice that were retro-orbitally injected with saline or ABE-AAV9 at P14.
Immunofluorescence staining of C57BL/6 (n = 2), untreated P28 homozygous LMNA c.1824 C>T (n = 1), saline-treated homozygous LMNA c.1824 C>T (n = 4) and ABE-treated homozygous LMNA c.1824 C>T (n = 5) mouse aortas stained for human lamin A/C + DAPI or for progerin + DAPI. Scale bars, 10 μm. Images from untreated 28 day-old, WT M1, 9424M and 9464M are replicated from Fig. 4 for ease of comparison.
Extended Data Fig. 9 Skin histology of male and female mice that were retro-orbitally injected with saline or ABE-AAV9 at P3.
a, Representative skin cross-sections for P3 RO saline-injected (left), ABE-AAV9-injected (middle) and wild-type C57BL/6 males at 6 months of age. Left images were stained with H&E; right images were prepared with Masson’s trichrome staining. b, Representative skin cross-sections for P3 RO saline-injected (left), ABE-AAV9-injected (middle) and wild-type C57BL/6 females at 6 months of age. Left images were stained with H&E; right images were prepared with Masson’s trichrome staining. Unaffected WT are wild-type C57BL/6 mice. These sections each represent replicates from different mice.
Extended Data Fig. 10 Skin histology of male and female mice that were retro-orbitally injected with saline or ABE-AAV9 at P14.
a, Representative skin cross-sections for P14 RO saline-injected (left), ABE-AAV9-treated (middle) and wild-type C57BL/6 males at 6 months of age. Left images were stained with H&E; right images were prepared with Masson’s trichrome staining. b, Representative skin cross-sections for P14 RO saline-injected (left), ABE-AAV9-treated (middle) and wild-type C57BL/6 females at 6 months of age. Left images were stained with H&E; right images were prepared with Masson’s trichrome staining. Unaffected WT are wild-type C57BL/6 mice, reproduced from Extended Data Fig. 12a, b for ease of comparison. These sections each represent replicates from different mice.
Extended Data Fig. 11 Body weights of mice that were retro-orbitally injected with saline or ABE-AAV9 at P3 or P14.
Weights of homozygous LMNA c.1824 C>T mice taken across mouse lifespans for cohorts of P3 RO (left) and P14 RO (right) saline- and ABE-AAV9-injected cohorts. Mouse weights are shown by sex. The x axis shows days post-injection, rather than age. Data are mean ± s.d. for the number of surviving mice at each time point; complete data can be accessed in Supplementary Data 5.
Extended Data Fig. 12 Whole-genome sequencing analysis of SNVs and indels and quantification of RAB25 transcript levels in mouse tissue samples.
a, Distribution of all possible SNV types in non-tumour liver tissue and liver tumour tissue samples isolated from ABE-AAV9-injected and saline-injected mice. Values from individual tissue samples are shown on the left. Aggregated values from all AAV-injected mouse tumour tissue samples, all AAV-injected mouse liver tissue samples and all saline-injected mouse liver tissue samples are shown on the right. Data are mean ± s.d. with each tissue section treated as a different sample: AAV-injected tumour tissue (n = 7), AAV-injected liver tissue (n = 6) and saline-injected liver tissue (n = 2). b, Genomic classification of A•T-to-G•C SNVs. Values from individual tissue samples are shown on the left. Aggregated values from AAV-injected mouse tumour tissue samples, AAV-injected mouse liver tissue samples and saline-injected mouse liver tissue samples from all tissue types are shown on the right. Data are mean ± s.d. with each tissue section treated as a different sample: AAV-injected tumour tissue (n = 7), AAV-injected liver tissue (n = 6), saline-injected liver tissue (n = 2). c, A•T-to-G•C SNVs and indels found in or near genes that are recurrently mutated in human liver cancers63,64 including introns, exons, and at ATAC-seq-defined cis-regulatory regions within 100-kb of each gene’s transcription start site, in AAV-injected mouse tumour tissue samples, AAV-injected mouse liver tissue samples and saline-injected mouse liver tissue samples. Data are mean ± s.d. Individual data points are shown for each sample. The complete list of SNVs from ANNOVAR analysis is provided as Supplementary Data 6. Summary statistics for SNV calls are in Supplementary Data 2. d, RNA isolated from mouse liver tissue samples was reverse-transcribed and amplified with primer sets specific to mouse Gapdh (detected with Cy5), mouse Actb (detected with Cy5.5) and human RAB25 (detected with TEX 615). Ct values were determined by quantitative PCR and are shown below each lane. N.D., not detected.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-3, Supplementary Figure 1, Supplementary Tables 1-2 and Supplementary References.
Supplementary Data
Supplementary Data 1: AAV integration analysis from mouse whole-genome sequencing data.
Supplementary Data
Supplementary Data 2: Analysis of SNVs from mouse whole-genome sequencing data.
Supplementary Data
Supplementary Data 3: Liver cancer-associated SNVs and indels in mouse wholegenome sequencing data.
Supplementary Data
Supplementary Data 4: CIRCLE-seq candidate off-target loci genomic coordinates and amplification primers.
Supplementary Data
Supplementary Data 5: Raw data for in vivo samples.
Supplementary Data
Supplementary Data 6: All SNVs from mouse whole-genome sequencing data.
Video 1
Representative untreated male progeria mouse (not enrolled in this study), taken at 7.5 months of age.
Video 2
Representative male progeria mice injected at P14 with ABE-AAV, taken at 9.75 months of age.
Video 3
Representative female progeria mice injected at P14 with ABE-AAV, and wild-type C57BL/6 female mouse, taken at 9.75 months of age.
Video 4
Representative male progeria mice injected at P14 with ABE-AAV, and wild-type C57BL/6 male mouse, taken at 11 months of age.
Video 5
Representative female progeria mice injected at P14 with ABE-AAV, taken at 11 months of age.
Rights and permissions
About this article
Cite this article
Koblan, L.W., Erdos, M.R., Wilson, C. et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature 589, 608–614 (2021). https://doi.org/10.1038/s41586-020-03086-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03086-7
This article is cited by
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Nature Communications (2024)
-
DNA and RNA base editors can correct the majority of pathogenic single nucleotide variants
npj Genomic Medicine (2024)
-
Base editing effectively prevents early-onset severe cardiomyopathy in Mybpc3 mutant mice
Cell Research (2024)
-
Whole-brain in vivo base editing reverses behavioral changes in Mef2c-mutant mice
Nature Neuroscience (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.