Abstract
Chronic liver disease due to alcohol-use disorder contributes markedly to the global burden of disease and mortality1,2,3. Alcoholic hepatitis is a severe and life-threatening form of alcohol-associated liver disease. The gut microbiota promotes ethanol-induced liver disease in mice4, but little is known about the microbial factors that are responsible for this process. Here we identify cytolysin—a two-subunit exotoxin that is secreted by Enterococcus faecalis5,6—as a cause of hepatocyte death and liver injury. Compared with non-alcoholic individuals or patients with alcohol-use disorder, patients with alcoholic hepatitis have increased faecal numbers of E. faecalis. The presence of cytolysin-positive (cytolytic) E. faecalis correlated with the severity of liver disease and with mortality in patients with alcoholic hepatitis. Using humanized mice that were colonized with bacteria from the faeces of patients with alcoholic hepatitis, we investigated the therapeutic effects of bacteriophages that target cytolytic E. faecalis. We found that these bacteriophages decrease cytolysin in the liver and abolish ethanol-induced liver disease in humanized mice. Our findings link cytolytic E. faecalis with more severe clinical outcomes and increased mortality in patients with alcoholic hepatitis. We show that bacteriophages can specifically target cytolytic E. faecalis, which provides a method for precisely editing the intestinal microbiota. A clinical trial with a larger cohort is required to validate the relevance of our findings in humans, and to test whether this therapeutic approach is effective for patients with alcoholic hepatitis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw 16S sequence reads can be found in the NCBI SRA associated with Bioproject PRJNA525701. Phage raw sequence reads and annotated genomes are available at NCBI under the following consecutive BioSample identifiers (SAMN11089809–SAMN11089827). Genome sequence data of E. faecalis strains isolated in this study were registered at the ENA under study PRJEB25007.
Code availability
The PERL script for making the genetic maps of phage genomes can be found at https://github.com/JCVenterInstitute/LinearDisplay.
References
Lozano, R. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012).
Lee, B. P., Vittinghoff, E., Dodge, J. L., Cullaro, G. & Terrault, N. A. National trends and long-term outcomes of liver transplant for alcohol-associated liver disease in the United States. JAMA Intern. Med. 179, 340–348 (2019).
Rehm, J. et al. Burden of disease associated with alcohol use disorders in the United States. Alcohol. Clin. Exp. Res. 38, 1068–1077 (2014).
Llopis, M. et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016).
Ike, Y., Clewell, D. B., Segarra, R. A. & Gilmore, M. S. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 172, 155–163 (1990).
Tang, W. & van der Donk, W. A. The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat. Chem. Biol. 9, 157–159 (2013).
Maddrey, W. C. et al. Corticosteroid therapy of alcoholic hepatitis. Gastroenterology 75, 193–199 (1978).
Dominguez, M. et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am. J. Gastroenterol. 103, 2747–2756 (2008).
Thursz, M. R. et al. Prednisolone or pentoxifylline for alcoholic hepatitis. N. Engl. J. Med. 372, 1619–1628 (2015).
Mathurin, P. & Lucey, M. R. Management of alcoholic hepatitis. J. Hepatol. 56, S39–S45 (2012).
Llorente, C. et al. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat. Commun. 8, 837 (2017).
Gilmore, M. S. et al. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176, 7335–7344 (1994).
Cox, C. R., Coburn, P. S. & Gilmore, M. S. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 6, 77–84 (2005).
Van Tyne, D., Martin, M. J. & Gilmore, M. S. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 5, 895–911 (2013).
Bertola, A., Mathews, S., Ki, S. H., Wang, H. & Gao, B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 8, 627–637 (2013).
Ogilvie, L. A. & Jones, B. V. The human gut virome: a multifaceted majority. Front. Microbiol. 6, 918 (2015).
Chatterjee, A. et al. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect. Immun. 87, e00085-19 (2019).
Poindexter, J. S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28, 231–295 (1964).
Shin, J. et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci. Rep. 6, 29681 (2016).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Marcuk, L. M. et al. Clinical studies of the use of bacteriophage in the treatment of cholera. Bull. World Health Organ. 45, 77–83 (1971).
Sarker, S. A. et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137 (2016).
Dalmasso, M., Hill, C. & Ross, R. P. Exploiting gut bacteriophages for human health. Trends Microbiol. 22, 399–405 (2014).
Ujmajuridze, A. et al. Adapted bacteriophages for treating urinary tract infections. Front. Microbiol. 9, 1832 (2018).
Khawaldeh, A. et al. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 60, 1697–1700 (2011).
Schooley, R. T. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954-17 (2017).
Dedrick, R. M. et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019).
Fish, R. et al. Compassionate use of bacteriophage therapy for foot ulcer treatment as an effective step for moving toward clinical trials. Methods Mol. Biol. 1693, 159–170 (2018).
Górski, A. et al. Phages and immunomodulation. Future Microbiol. 12, 905–914 (2017).
Brandl, K. et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 69, 396–405 (2018).
Gao, B. et al. Serum and fecal oxylipins in patients with alcohol-related liver disease. Dig. Dis. Sci. 64, 1878–1892 (2019).
Lang, S. et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology (2019).
Ball, S. A., Tennen, H., Poling, J. C., Kranzler, H. R. & Rounsaville, B. J. Personality, temperament, and character dimensions and the DSM-IV personality disorders in substance abusers. J. Abnorm. Psychol. 106, 545–553 (1997).
Krieg, L. et al. Mutation of the gastric hydrogen-potassium ATPase alpha subunit causes iron-deficiency anemia in mice. Blood 118, 6418–6425 (2011).
Gill, J. J. et al. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13, 542 (2012).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3, e000132 (2017).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Santiago-Rodriguez, T. M. et al. Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genomics 16, 549 (2015).
Haft, D. H. et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860 (2018).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016).
Fouts, D. E. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34, 5839–5851 (2006).
Ondov, B. D. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Valentine, R. C., Shapiro, B. M. & Stadtman, E. R. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7, 2143–2152 (1968).
Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108 (Suppl 1), 4516–4522 (2011).
Chen, P. et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology 148, 203–214.e16 (2015).
Ryu, H. et al. Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of enterococcus species in environmental samples. Appl. Environ. Microbiol. 79, 196–204 (2013).
Haas, W., Shepard, B. D. & Gilmore, M. S. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87 (2002).
Page, A. J. et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb. Genom. 2, e000083 (2016).
Chen, L. et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328 (2005).
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017)
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Tang, W., Bobeica, S. C., Wang, L. & van der Donk, W. A. CylA is a sequence-specific protease involved in toxin biosynthesis. J. Ind. Microbiol. Biotechnol. 46, 537–549 (2019).
Iwaisako, K. et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor δ agonist. Proc. Natl Acad. Sci. USA 109, E1369–E1376 (2012).
Clarke, T. H., Brinkac, L. M., Sutton, G. & Fouts, D. E. GGRaSP: a R-package for selecting representative genomes using Gaussian mixture models. Bioinformatics 34, 3032–3034 (2018).
Acknowledgements
S. Lang was supported by a DFG fellowship (LA 4286/1-1), C.L. was supported by an AASLD Pinnacle Research Award in Liver Disease and a pilot project award from Southern California Research Center for Alcoholic Liver and Pancreatic Disease (ALPD) and Cirrhosis (P50 AA011999), Y.S. was supported by a Wellcome Trust PhD studentship and I.R.R. was supported by National Institute of General Medical Sciences (NIGMS)–NIH Chemistry–Biology Interface Training Grant (T32-GM070421). This study was supported in part by a Biocodex Microbiota Foundation Grant, NIH grants R01 AA24726, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.), the Wellcome Trust (WT098051) (to T.D.L.), NIH grant U01AA021908 (to R.B.) and services provided by P30 DK120515 and P50 AA011999.
Author information
Authors and Affiliations
Contributions
Y.D. was responsible for acquisition, analysis and interpretation of data, and drafting of the manuscript; C.L. was responsible for study concept and design, acquisition, analysis and interpretation of data and key preliminary experiments; S. Lang, K.B., J.L. and X.M.T. provided assistance with statistical analysis; H.C., L.J., B.G., W.S., R.K., F.H. and S. Lee provided assistance with data acquisition; R.C.W., T.H.C., K.N., M.T. and D.E.F. were responsible for 16S rRNA sequencing, phage genome sequencing and data analysis; Y.S. and T.D.L. were responsible for bacterial genome sequencing and data analysis; A.H.-M., L.L. and R.Y. provided assistance with phage studies and were responsible for electron microscopy data; R.Y. provided critical revision of the manuscript; I.R.R. and W.A.v.d.D. were responsible for cytolysin expression and purification; Y.M. and L.E. provided assistance with the design and conduct of the gnotobiotic mouse studies; M.L. and D.P. provided assistance with phage isolation; M.V.-C., F.B.-P., E.C.V., J.G.A., R.S.B. Jr, V.V., J.A., J.C., D.L.S., S.B.H., A.L., M.R.L., P.M., G.G.-T., R.B. and P.S. were responsible for collection of human samples; D.E.F. and B.S. were responsible for the study concept and design, and editing the manuscript; B.S. was responsible for study supervision.
Corresponding author
Ethics declarations
Competing interests
B.S. consults for the Ferring Research Institute; however, there is no competing interest with regard to this study. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Intestinal dysbiosis in patients with alcoholic hepatitis.
a, 16S rRNA sequencing of faecal samples from controls (n = 14), patients with alcohol-use disorder (n = 43), or alcoholic hepatitis (n = 75). The graph demonstrates the relative abundance of sequence reads in each genus. b, Bacterial diversity (Shannon index and Simpson index) and richness (Chao richness) was calculated in controls (n = 14), patients with alcohol-use disorder (n = 43) or alcoholic hepatitis (n = 75). c, E. faecalis in faecal samples from controls (n = 25), patients with alcohol-use disorder (n = 38) or alcoholic hepatitis (n = 82), assessed by qPCR. d, Percentage of faecal samples positive for E. faecalis in controls (n = 25), patients with alcohol-use disorder (n = 38) or alcoholic hepatitis (n = 82), assessed by qPCR. E. faecalis was detected in faeces from 80% of patients with alcoholic hepatitis, versus 36% of controls (P < 0.001). There was also a significant difference between patients with alcohol-use disorder and patients with alcoholic hepatitis (P < 0.01). e, Receiver operating characteristic curves and area under the curve (AUC) for the comparison of 90-day mortality and cytolysin positivity (red; n = 57), MELD score (blue; n = 56), ABIC score (yellow; n = 57) and discriminant function (green; n = 42) in patients with alcoholic hepatitis. f, E. faecalis in faecal samples from patients with alcoholic hepatitis whose faecal samples were cytolysin-positive (n = 25) or cytolysin-negative (n = 54), assessed by qPCR (P = 0.8174). g, 16S rRNA sequencing of faecal samples from patients with alcoholic hepatitis from different centres (France, n = 9; Mexico, n = 6; Spain, n = 5; UK, n = 11; USA (east), n = 16; USA (Midwest), n = 12; USA (west), n = 16 patients). We used PCoA based on Jaccard dissimilarity matrices to show β-diversity among groups at the genus level. The composition of faecal microbiota was significantly different between patients from different regions (P < 0.01). h, Percentage of faecal samples that were positive for cylLL and cylLS DNA sequences (cytolysin-positive), in patients with alcoholic hepatitis from different centres (France, n = 16; Mexico, n = 6; Spain, n = 6; UK, n = 10; USA (east), n = 16; USA (Midwest), n = 13; USA (west), n = 15 patients), assessed by qPCR (P = 0.6094). i, E. faecalis in faecal samples from patients with alcoholic hepatitis from different centres, assessed by qPCR (P = 0.5648). j, Percentage of faecal samples that were positive for E. faecalis in patients with alcoholic hepatitis from different centres (France, n = 16; Mexico, n = 6; Spain, n = 6; UK, n = 10; USA (east), n = 16; USA (Midwest), n = 13; USA (west), n = 15 patients), assessed by qPCR (P = 0.0529). k, Percentage of subjects with faecal samples that were positive for cylLL and cylLS DNA sequences (cytolysin-positive), in patients with alcoholic hepatitis and with (n = 30) or without (n = 18) cirrhosis, assessed by qPCR (P = 0.3431). l, E. faecalis in faecal samples from patients with alcoholic hepatitis and with (n = 30) or without (n = 18) cirrhosis, assessed by qPCR (P = 0.5736). m, Percentage of faecal samples that were positive for E. faecalis in patients with alcoholic hepatitis and with (n = 30) or without (n = 18) cirrhosis, assessed by qPCR (P = 0.2878). Results are expressed as mean ± s.e.m. (c, f, i, l). For the box and whisker plots in b, the box extends from the 25th to 75th percentiles, and the centre line represents the median; for all three groups, the bottom whiskers show the minimum values; for the control group (black), the top whisker shows the maximum value; for the other two groups, the top whiskers represent the 75th percentile plus 1.5× the inter-quartile distance (the distance between the 25th and 75th percentiles); all values greater than this are plotted as individual dots. P values were determined by Kruskal–Wallis test (i) with Dunn’s post hoc test (b, c), two-sided Fisher’s exact test (h, j, k, m) followed by FDR procedures (d), two-sided Mann–Whitney Wilcoxon rank-sum test (f, l) or PERMANOVA (g). The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 2 Cytolytic E. faecalis causes the progression of ethanol-induced liver disease in mice.
a–n, C57BL/6 mice were fed oral isocaloric (control) or chronic–binge ethanol diets and gavaged with vehicle (PBS), a cytolytic E. faecalis strain (FA2-2(pAM714)) (denoted E. faecalis) (5 × 108 CFUs) or a non-cytolytic E. faecalis strain (FA2-2(pAM771))5 (denoted E. faecalis Δcytolysin) (5 × 108 CFUs) every third day. a, Serum levels of ALT. b, Hepatic triglyceride content. c, Representative oil red O-stained liver sections. d–f, Hepatic levels of mRNAs. g, Kaplan–Meier curve of survival of mice on chronic–binge ethanol diets (day 0 denotes the start of ethanol feeding). Mice gavaged with PBS all survived, and are not included in the figure. A higher proportion of mice (n = 15) gavaged with non-cytolytic E. faecalis survived than did mice (n = 25) gavaged with cytolytic E. faecalis. h, Proportions of mice that were positive for cytolysin in the liver, measured by qPCR for cylLS (the gene that encodes cytolysin subunit CylLS′′). i, Proportions of mice that were positive for E. faecalis in the liver, measured by qPCR. About 80% of mice colonized with cytolytic E. faecalis, as well as those colonized with non-cytolytic E. faecalis, were positive for E. faecalis in their livers. j, Liver CFUs of Enterococcus in mice on a chronic–binge ethanol diet. k, Paracellular intestinal permeability was evaluated by measuring faecal albumin content and serum levels of lipopolysaccharide (LPS) by enzyme-linked immunosorbent assays. l, Faecal samples were collected and 16S rRNA genes were sequenced. PCoA based on Jaccard dissimilarity matrices showed no significant differences among mice gavaged with PBS, cytolytic or non-cytolytic E. faecalis following feeding with the control and ethanol diets. Compared to mice fed with a control diet, mice fed with an ethanol diet had significantly different faecal microbiomes after gavaging with E. faecalis (P < 0.05). m, n, Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among mice gavaged with PBS, cytolytic or non-cytolytic E. faecalis after ethanol feeding. o, Mice were gavaged with cytolytic or non-cytolytic E. faecalis strains (carrying the erythromycin resistance gene; 5 × 108 CFUs) at time 0, and faeces were collected 0, 8, 24, 48 and 72 h later. Faecal CFUs of Enterococcus were determined by culturing faecal samples on BBL enterococcosel broth agar plate with 50 μg ml-1 erythromycin. At time 0 and 72 h, five out of five and four out of five mice, respectively, had no detectable erythromycin-resistant Enterococcus in their faeces. These points are not shown on the graph, but have been included in the calculation of mean ± s.e.m. Scale bar, 100 μm. Results are expressed as mean ± s.e.m. (a, b, d–f, j, k, m–o). P values among groups of mice fed with the control or ethanol diet were determined by one-way ANOVA with Tukey’s post hoc test (a, b, d–f, j, k, m, n), two-sided log-rank (Mantel–Cox) test (g), two-sided Fisher’s exact test followed by FDR procedures (h, i) or PERMANOVA followed by FDR procedures (l). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. P values between mice fed with a control diet and mice fed with an ethanol diet were determined by two-way ANOVA (k). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
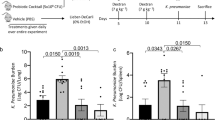
Extended Data Fig. 3 Transplantation of cytolysin-positive faeces increases ethanol-induced liver disease in gnotobiotic mice.
a–f, h, i, C57BL/6 germ-free mice were colonized with faeces from two cytolysin-positive and two cytolysin-negative patients with alcoholic hepatitis, and then fed isocaloric (control) or chronic–binge ethanol diets. a, Percentage of TUNEL-positive hepatic cells. b, Representative oil red O-stained liver sections. c, d, Hepatic levels of mRNAs that encode the inflammatory cytokine Cxcl2 and Acta2 (a marker of activated hepatic stellate cells). e, Kaplan–Meier curve of survival of mice on chronic–binge ethanol diets (day 0 denotes the start of ethanol feeding), gavaged with faeces from cytolysin-positive (n = 48 mice) or cytolysin-negative (n = 32 mice) patients with alcoholic hepatitis. f, Faecal samples were collected and 16S rRNA genes were sequenced. The graph shows PCoA of faecal microbiomes. No significant difference was observed between mice colonized with faeces from cytolysin-positive or cytolysin-negative donors with alcoholic hepatitis, following the control diet. Mice transplanted with faeces from a cytolysin-positive patient with alcoholic hepatitis (patient no. 2) showed a microbiota that was significantly different to that of the other mouse groups following ethanol administration (P < 0.01). g, Percentage of cytolysin-positive E. faecalis in four patients with alcoholic hepatitis. Stool samples from the four patients were placed on plates with selective medium, and Enterococcus colonies were identified by the production of a dark brown or black colour. Enterococcus colonies were confirmed to be E. faecalis by qPCR. The cytolysin status of each E. faecalis colony was determined by qPCR. h, Serum levels of ethanol were comparable among colonized mice after ethanol feeding. i, Hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice on control or ethanol diets. Scale bar, 100 μm. Results are expressed as mean ± s.e.m. (a, c, d, h, i). P values were determined by one-way ANOVA with Tukey’s post hoc test (a, c, d, h, i), two-sided log-rank (Mantel–Cox) test (e) or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Fig. 4 Isolation and amplification of phages against cytolytic E. faecalis isolated from mice.
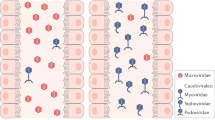
a, BHI agar plate showing phage plaque morphology. The phage cocktail (100 μl) (102–103 PFUs) was mixed with overnight-grown E. faecalis culture (100 μl) and then added to BHI broth top agar (0.5% agar) and poured over a BHI plate (1.5% agar). After overnight growth at 37 °C, images were captured on an Epson Perfection 4990 Photo scanner. b, Simplified illustration of the morphologies of different phages. Siphophages have long, flexible noncontractile tails (left); myophages have contractile tails (middle); and podophages have short noncontractile tails (right). c, Transmission electron microscopy revealed that phages we isolated were all podophages (Efmus1, Efmus2, Efmus3 and Efmus4). d, Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (in bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are coloured on the basis of functional role categories. Scale bar, 50 nm. All results were generated from at least three independent replicates.
Extended Data Fig. 5 Phages reduce translocation of cytolysin to the liver and reduce ethanol-induced liver disease in Atp4aSl/Sl mice.
a–k, Wild-type (WT) and Atp4aSl/Sl littermates were fed oral isocaloric (control) or chronic–binge ethanol diets, and gavaged with vehicle (PBS), control phages against C. crescentus (1010 PFUs) or a cocktail of four different phages that target cytolytic E. faecalis (1010 PFUs), 1 day before an ethanol binge. a, Serum levels of ALT. b, Hepatic triglyceride content. c, Representative oil red O-stained liver sections. d–f, Hepatic levels of mRNAs. g, Proportions of mice that were positive for cytolysin in the liver, measured by qPCR for cylLS. h, Faecal CFUs of Enterococcus. i, Faecal samples were collected and 16S rRNA genes were sequenced. PCoA based on Jaccard dissimilarity matrices found no significant difference in faecal microbiota among mice given PBS, control phage or phages that target cytolytic E. faecalis in each group. j, k, Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar, 100 μm. Results are expressed as mean ± s.e.m. (a, b, d–f, h, j, k). P values were determined by two-way ANOVA with Tukey’s post hoc test (a, b, d–f, h, j, k), two-sided Fisher’s exact test followed by FDR procedures (g) or PERMANOVA followed by FDR procedures (i). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 6 Isolation and amplification of phages against cytolytic E. faecalis strains isolated from patients with alcoholic hepatitis.
a, BHI agar plates showing phage plaque morphology. b, Transmission electron microscopy graphs of myophages Ef2.1 and Ef2.3, stained with phosphotungstic acid showing contracted tails. c, Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (in bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are coloured on the basis of functional role categories. Sequences that encode tRNA genes are indicated by a cloverleaf structure. Scale bar, 50 nm. All results were generated from at least three independent replicates.
Extended Data Fig. 7 Phages that target cytolytic E. faecalis reduce ethanol-induced liver disease in gnotobiotic mice.
a–h, C57BL/6 germ-free mice were colonized with faeces from two cytolysin-positive patients with alcoholic hepatitis (faeces from one patient were also used in Fig. 2). The mice were then fed oral isocaloric (control) or chronic–binge ethanol diets, and gavaged with control phages against C. crescentus (1010 PFUs) or a cocktail of 3 or 4 different phages that target cytolytic E. faecalis (1010 PFUs), one day before an ethanol binge. a, Percentage of TUNEL-positive hepatic cells. b, Representative oil red O-stained liver sections. c, d, Hepatic levels of mRNAs that encode the inflammatory cytokine Cxcl2, and Acta2 (a marker of activated hepatic stellate cells). e, Faecal CFUs of Enterococcus. f, Faecal samples were collected and 16S rRNA genes were sequenced. PCoA based on Jaccard dissimilarity matrices shows no significant differences in the faecal microbiota of mice gavaged with control phage and phages that target cytolytic E. faecalis in each group. g, h, Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar, 100 μm. Results are expressed as mean ± s.e.m. (a, c–e, g, h). P values were determined by two-way ANOVA with Tukey’s post hoc test (a, c–e, g, h) or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P < 0.05, ***P < 0.001.
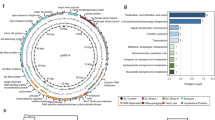
Extended Data Fig. 8 Isolation and amplification of phages against non-cytolytic E. faecalis strains isolated from patients with alcoholic hepatitis.
a, BHI agar plates showing phage plaque morphology. b, Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (in bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are coloured on the basis of functional role categories. Sequences that encode tRNA genes are indicated by a cloverleaf structure. c, Phylogenetic tree of Enterococcus phages. A whole-genome average nucleotide distance tree was constructed for 73 available Enterococcus phage genomes: 54 of these were from GenBank (denoted by black letters) and 19 were from this study (4 phages against cytolysin-positive E. faecalis isolated from mice (shown in blue letters); 7 phages against cytolysin-positive E. faecalis isolated from patients with alcoholic hepatitis (shown in pink letters); and 8 phages against cytolysin-negative E. faecalis isolated from patients with alcoholic hepatitis (shown in green letters)) with Mash43 using a sketch size of s = 5000 and a k-mer size of k = 12 and GGRaSP59 (Methods). Coloured branches denote specific phage genera or subfamily: Sap6virus, P68virus and Spounavirinae. The scale bar represents per cent average nucleotide divergence. All results were generated from at least three independent replicates.
Extended Data Fig. 9 Phages that target non-cytolytic E. faecalis do not reduce ethanol-induced liver disease in gnotobiotic mice.
a–h, C57BL/6 germ-free mice were colonized with faeces from two cytolysin-negative patients with alcoholic hepatitis. Transplanted gnotobiotic mice were fed oral isocaloric (control) or chronic–binge ethanol diets and gavaged with control phages against C. crescentus (1010 PFUs) or a cocktail of four different phages targeting non-cytolytic E. faecalis (1010 PFUs), 1 day before an ethanol binge. a, Percentage of TUNEL-positive hepatic cells. b, Representative oil red O-stained liver sections. c, d, Hepatic levels of mRNAs that encode the inflammatory cytokine Cxcl2, and Acta2 (a marker of activated hepatic stellate cells). e, Proportions of mice that were positive for cytolysin in the liver, measured by qPCR for cylLS. f, Faecal samples were collected and 16S rRNA genes were sequenced. PCoA based on Jaccard dissimilarity matrices found no significant difference in faecal microbiota among mice gavaged with control phages and phages that target cytolytic E. faecalis in each group. g, h, Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar, 100 μm. Results are expressed as mean ± s.e.m. (a, c, d, g, h). P values were determined by two-way ANOVA with Tukey’s post hoc test (a, c, d, g, h), two-sided Fisher’s exact test followed by FDR procedures (e) or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-9.
Supplementary Table
This file contains Supplementary Table 10.
Rights and permissions
About this article
Cite this article
Duan, Y., Llorente, C., Lang, S. et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019). https://doi.org/10.1038/s41586-019-1742-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1742-x
This article is cited by
-
The gut-liver axis in hepatobiliary diseases
Inflammation and Regeneration (2024)
-
Utilization of the microbiome in personalized medicine
Nature Reviews Microbiology (2024)
-
Phage-specific immunity impairs efficacy of bacteriophage targeting Vancomycin Resistant Enterococcus in a murine model
Nature Communications (2024)
-
Integrative metagenomic and metabolomic analyses reveal the potential of gut microbiota to exacerbate acute pancreatitis
npj Biofilms and Microbiomes (2024)
-
Phage cocktail alleviated type 2 diabetes by reshaping gut microbiota and decreasing proinflammatory cytokines
Applied Microbiology and Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.