Abstract
High-grade gliomas are lethal brain cancers whose progression is robustly regulated by neuronal activity. Activity-regulated release of growth factors promotes glioma growth, but this alone is insufficient to explain the effect that neuronal activity exerts on glioma progression. Here we show that neuron and glioma interactions include electrochemical communication through bona fide AMPA receptor-dependent neuron–glioma synapses. Neuronal activity also evokes non-synaptic activity-dependent potassium currents that are amplified by gap junction-mediated tumour interconnections, forming an electrically coupled network. Depolarization of glioma membranes assessed by in vivo optogenetics promotes proliferation, whereas pharmacologically or genetically blocking electrochemical signalling inhibits the growth of glioma xenografts and extends mouse survival. Emphasizing the positive feedback mechanisms by which gliomas increase neuronal excitability and thus activity-regulated glioma growth, human intraoperative electrocorticography demonstrates increased cortical excitability in the glioma-infiltrated brain. Together, these findings indicate that synaptic and electrical integration into neural circuits promotes glioma progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA sequencing of single-cell patient derived xenografts is available on Gene Expression Omnibus (GEO) under accession GSE134269. All other data are available in the article, source data, or from the corresponding author upon reasonable request.
Code availability
Custom code created will be available on GitHub upon request.
References
Venkatesh, H. S. et al. Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell 161, 803–816 (2015).
Venkatesh, H. S. et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549, 533–537 (2017).
Bergles, D. E., Roberts, J. D., Somogyi, P. & Jahr, C. E. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191 (2000).
Káradóttir, R., Cavelier, P., Bergersen, L. H. & Attwell, D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438, 1162–1166 (2005).
LoTurco, J. J., Owens, D. F., Heath, M. J., Davis, M. B. & Kriegstein, A. R. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 15, 1287–1298 (1995).
Luk, K. C. & Sadikot, A. F. Glutamate and regulation of proliferation in the developing mammalian telencephalon. Dev. Neurosci. 26, 218–228 (2004).
Liu, X., Wang, Q., Haydar, T. F. & Bordey, A. Nonsynaptic GABA signaling in postnatal subventricular zone controls proliferation of GFAP-expressing progenitors. Nat. Neurosci. 8, 1179–1187 (2005).
Deisseroth, K. et al. Excitation–neurogenesis coupling in adult neural stem/progenitor cells. Neuron 42, 535–552 (2004).
Kougioumtzidou, E. et al. Signalling through AMPA receptors on oligodendrocyte precursors promotes myelination by enhancing oligodendrocyte survival. eLife 6, e28080 (2017).
Filbin, M. G. et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335 (2018).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Sommer, B., Köhler, M., Sprengel, R. & Seeburg, P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 (1991).
Hollmann, M., Hartley, M. & Heinemann, S. Ca2+ permeability of KA-AMPA-gated glutamate receptor channels depends on subunit composition. Science 252, 851–853 (1991).
Venkataramani, V. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature https://doi.org/10.1038/s41586-019-1564-x (2019).
Oliet, S. H., Malenka, R. C. & Nicoll, R. A. Bidirectional control of quantal size by synaptic activity in the hippocampus. Science 271, 1294–1297 (1996).
Bergles, D. E. & Jahr, C. E. Synaptic activation of glutamate transporters in hippocampal astrocytes. Neuron 19, 1297–1308 (1997).
Lüscher, C., Malenka, R. C. & Nicoll, R. A. Monitoring glutamate release during LTP with glial transporter currents. Neuron 21, 435–441 (1998).
Sibille, J., Pannasch, U. & Rouach, N. Astroglial potassium clearance contributes to short-term plasticity of synaptically evoked currents at the tripartite synapse. J. Physiol. 592, 87–102 (2014).
McKhann, G. M. II, D’Ambrosio, R. & Janigro, D. Heterogeneity of astrocyte resting membrane potentials and intercellular coupling revealed by whole-cell and gramicidin-perforated patch recordings from cultured neocortical and hippocampal slice astrocytes. J. Neurosci. 17, 6850–6863 (1997).
Osswald, M. et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015).
Labrakakis, C., Patt, S., Hartmann, J. & Kettenmann, H. Glutamate receptor activation can trigger electrical activity in human glioma cells. Eur. J. Neurosci. 10, 2153–2162 (1998).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Ishiuchi, S. et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J. Neurosci. 27, 7987–8001 (2007).
Sontheimer, H. A role for glutamate in growth and invasion of primary brain tumors. J. Neurochem. 105, 287–295 (2008).
Lyons, S. A., Chung, W. J., Weaver, A. K., Ogunrinu, T. & Sontheimer, H. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res. 67, 9463–9471 (2007).
Chen, Q. et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016).
Campbell, S. L., Buckingham, S. C. & Sontheimer, H. Human glioma cells induce hyperexcitability in cortical networks. Epilepsia 53, 1360–1370 (2012).
John Lin, C. C. et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405 (2017).
Buckingham, S. C. et al. Glutamate release by primary brain tumors induces epileptic activity. Nat. Med. 17, 1269–1274 (2011).
Campbell, S. L. et al. GABAergic disinhibition and impaired KCC2 cotransporter activity underlie tumor-associated epilepsy. Glia 63, 23–36 (2015).
Ray, S., Crone, N. E., Niebur, E., Franaszczuk, P. J. & Hsiao, S. S. Neural correlates of high-gamma oscillations (60-200 Hz) in macaque local field potentials and their potential implications in electrocorticography. J. Neurosci. 28, 11526–11536 (2008).
Yizhar, O. et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011).
Hodgkin, A. L. & Katz, B. The effect of sodium ions on the electrical activity of giant axon of the squid. J. Physiol. 108, 37–77 (1949).
Ransom, B. R. & Goldring, S. Ionic determinants of membrane potential of cells presumed to be glia in cerebral cortex of cat. J. Neurophysiol. 36, 855–868 (1973).
Bittman, K. S. & LoTurco, J. J. Differential regulation of connexin 26 and 43 in murine neocortical precursors. Cereb. Cortex 9, 188–195 (1999).
LoTurco, J. J., Blanton, M. G. & Kriegstein, A. R. Initial expression and endogenous activation of NMDA channels in early neocortical development. J. Neurosci. 11, 792–799 (1991).
Marins, M. et al. Gap junctions are involved in cell migration in the early postnatal subventricular zone. Dev. Neurobiol. 69, 715–730 (2009).
Ohtaka-Maruyama, C. et al. Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science 360, 313–317 (2018).
Kuffler, S. W. Neuroglial cells: physiological properties and a potassium mediated effect of neuronal activity on the glial membrane potential. Proc. R. Soc. Lond. B 168, 1–21 (1967).
Yizhar, O., Fenno, L. E., Davidson, T. J., Mogri, M. & Deisseroth, K. Optogenetics in neural systems. Neuron 71, 9–34 (2011).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Lin, G. L. et al. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol. Commun. 6, 51 (2018).
Qin, E. Y. et al. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell 170, 845–859.e819 (2017).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Pietzsch, T., Preibisch, S., Tomancák, P. & Saalfeld, S. ImgLib2—generic image processing in Java. Bioinformatics 28, 3009–3011 (2012).
Wu, D. et al. Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature 544, 316–321 (2017).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Achanta, R. et al. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34, 2274–2282 (2012).
Pnevmatikakis, E. A. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
van der Walt, S. et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014).
Kawahara, Y., Ito, K., Sun, H., Kanazawa, I. & Kwak, S. Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur. J. Neurosci. 18, 23–33 (2003).
Sobolevsky, A. I., Rosconi, M. P. & Gouaux, E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009).
Vinci, M., Box, C. & Eccles, S. A. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp. 99, 52686 (2015).
Vinci, M., Box, C., Zimmermann, M. & Eccles, S. A. Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol. Biol. 986, 253–266 (2013).
Acknowledgements
We gratefully acknowledge support from the National Institutes of Health (NIH) Director’s Common Fund (DP1 NS111132 to M.M.), National Institute of Neurological Disorders and Stroke (R01 NS092597 to M.M., K08 NS110919 to S.H.-J.), National Cancer Institute (F31 CA200273 to H.S.V.), National Institutes of Mental Health (P50 MH086403 to R.C.M.), Michael Mosier Defeat DIPG Foundation (to M.M.), ChadTough Foundation (to M.M.), V Foundation (to M.M.), Department of Defense (NF140075 to M.M.), McKenna Claire Foundation (to M.M.), Alex’s Lemonade Stand Foundation (to M.M.), The Cure Starts Now Foundation and DIPG Collaborative (to M.M.), Unravel Pediatric Cancer (to M.M.), N8 Foundation (to M.M.), Abbie’s Army Foundation (to M.M.), Brantley’s Project supported by Ian’s Friends Foundation (to M.M.), Waxman Family Research Fund (to M.M.), Joey Fabus Childhood Cancer Foundation (to M.M.), Virginia and D.K. Ludwig Fund for Cancer Research (to M.M.), Bio-X Institute (to L.T.T. and A.C.G.), Maternal and Child Health Research Institute at Stanford (to M.M., A.C.G and H.S.V.), Anne T. and Robert M. Bass Endowed Faculty Scholarship in Pediatric Cancer and Blood Diseases (to M.M.), Cancer Research UK (to M.M.), Dr. Mildred Scheel Cancer Foundation (57406718 to M.A.), Damon Runyan Foundation (to K.R.T.), Sontag Foundation Distinguished Scientist Award (to M.L.S.), Howard Hughes Medical Institute (to A.R.), Klarman Cell Observatory (to A.R.), Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to D.E.B.), Chica and Heinz Schaller Research Foundation (to A.A.), Deutsche Forschungsgemeinschaft (AG 287/1-1 to A.A.), The Robert Wood Johnson Foundation (74259 RWJF to S.H.-J.). We thank S. Knemeyer for illustrations, and A. Olsen, S. Kakaizada and M. Shore for technical assistance.
Author information
Authors and Affiliations
Contributions
H.S.V. and M.M. designed, conducted, and analysed experiments. W.M. conducted electrophysiology experiments. L.N. and H.V. contributed to electron microscopy data acquisition and analyses. S.H.-J. performed intraoperative electrocorticography. D.B. conducted high-gamma frequency power computational analyses. D.S., M.L.S., A.R. and S.M.G. contributed to single-cell transcriptomic analyses. S.M.G. and M.A. contributed to synaptic puncta and tumour microtube confocal imaging, A.C.G. and L.T.T. contributed to optogenetic experiments. A.A. and D.E.B. provided the GluA2 dominant-negative construct. A.P., K.R.T. and P.J.W. contributed to in vitro and in vivo data collection and analyses. C.E. contributed to quantitative imaging analyses. R.C.M., D.E.B., S.M.G., W.M., H.S.V. and M.M. contributed to manuscript editing. H.S.V. and M.M. wrote the manuscript. M.M. conceived of the project and supervised all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
M.M. is an SAB member of Cygnal Therapeutics. A.R. is a founder and equity holder of Celsius Therapeutics and an SAB member of ThermoFisher Scientific, Neogene Therapeutics and Syros Pharmaceuticals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Andres Barria, Michael Taylor and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Synaptic gene expression in single cell primary glioma and patient-derived xenografts.
a, Primary human biopsy single-cell transcriptomic data illustrating synapse associated and ion channel gene expression in H3K27M+ DMG (grey, n = 2,259 cells, 6 study participants), wild-type IDH adult high-grade glioma (red, n = 599 cells, 3 participants), IDH-mutant adult high-grade glioma (purple, n = 5,096 cells, 10 particpants) malignant cells, and tumour-associated, non-malignant immune cells (green; n = 96 cells, 5 participants) and oligodendrocytes (yellow; n = 232 cells). b, As in a for single-cell transcriptomic analysis of H3K27M+ DMG xenograft models (SU-DIPG-VI, blue; and SU-DIPG-XIII-FL, yellow) illustrating broad synaptic gene expression similar to that found in primary DMG tissue samples as shown in Fig. 1a. For each individual violin plot, the y axis represents log2(transcripts per million), and the x axis represents number of individual cells with indicated expression value.
Extended Data Fig. 2 Synaptic gene expression and structural synapses in glioma.
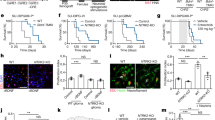
a, Plot of the lineage (x axis) and stemness (y axis) scores for H3K27M+ DIPG malignant single cells (dots) sorted from patient-derived glioma xenograft models (SU-DIPG-VI and SU-DIPG-XIII-FL). Overlay of grey–red enrichment score indicates relative score for OPC-like genes (left) and synapse-related genes (right; n = 335 cells, 4 mice; Pearson correlation rho = 0.57, P < 0.0001). b, Plot of the lineage versus stemness scores for H3K27M+ DIPG malignant single cells in a, coloured by xenograft model. Red denotes SU-DIPG-XIII-FL, blue denotes SU-DIPG-VI. Although each xenograft model clearly contains cells across all lineages, SU-DIPG-XIII-FL has a prominent astrocyte-like cell population, whereas SU-DIPG-VI has a prominent OPC-like population. c, Unbiased principal component analysis of single-cell gene expression from an individual patient primary biopsy sample of H3K27M+ glioma (BCH869) reveals cellular clusters that resemble proliferating cells (cell-cycle), OPCs (OPC-like), astrocytes (AC-like) and oligodendrocytes (OL-like). Synaptic gene enrichment for individual cells shown below and co-localizes in the OPC-like cluster. d, Electron microscopy images of primary adult glioblastoma tumour sample illustrating clear synaptic structure. Scale bars, 500 µm. e, Original (non-pseudo-coloured) immuno-electron microscopy images shown in Fig. 1c. f, Additional examples of neuron–glioma synapses identified by immuno-electron microscopy in patient-derived xenografts of SU-DIPG-VI and SU-DIPG-XIII-FL (n = 3 mice per group). Arrowheads indicate immuno-gold particle labelling of GFP. Scale bars, 200 µm. g, Quantification of neuron–glioma synaptic structures in SU-DIPG-VI (n = 101 sections) and SU-DIPG-XIII-FL (n = 104 sections) xenografts. For each individual violin plot, y axis represents number of identified unambiguous neuron–glioma synapses in each section, x axis represents number of individual cells with indicated value. h, Quantification of colocalized postsynaptic glioma-derived PSD95–RFP with presynaptic synapsin in co-cultures of SU-DIPG-XIII-FL glioma cells with wild-type or Nlgn3KO neurons (n = 21 cells, 10 coverslips per group). Data are mean ± s.e.m. ****P < 0.0001, two-sided Mann–Whitney test.
Extended Data Fig. 3 Mitogenic effects of NLGN3 are independent from AMPAR signalling and properties of glioma AMPARs.
a, Proliferative response of GFP (control) and GluA2-DN-expressing glioma cells after 24 h exposure to soluble extracellular NLGN3 (100 nM) in the presence and absence of AMPAR blocker NBQX (10 µM). b, Left, western blot analysis of phosphorylated-AKT (Ser473; pAKT) and total AKT in GFP (control) glioma cells in response to 5-min exposure to NLGN3 (100 nM) in the presence and absence of 10 µM NBQX. Right, quantitative analysis of the ratio of pAKT/AKT normalized to vehicle. c, Left, western blot analysis of pAKT and total AKT in GluA2-DN-expressing glioma cells in response to 5-min exposure to 100 nM NLGN3. Right, quantitative analysis of the ratio of pAKT/AKT normalized to vehicle. d, Left, time course of evoked glioma cell EPSC block by 100 µM NASPM (red bar denotes duration; n = 7 cells, 5 mice). Right, representative trace before (black) and after (red) addition of NASPM. e, Quantification of data in d. f, Q/R editing efficiency of GluA2 subunit in SU-DIPG-XIII-FL and SU-DIPG-VI cells as measured by PCR and expressed as percentage edited. g, Expression of ADAR1 (also known as ADAR), which encodes the enzyme responsible for Q/R editing of GluA2 (also known as GRIA2) mRNA. mRNA expression shown relative to ACTB and measured by quantitative PCR. Analyses in a–c, f, g were calculated from three independent sets of cells. Data are mean ± s.e.m. *P < 0.01, **P < 0.001, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Tukey’s post hoc analysis (a, b), two-tailed unpaired Student’s t-test (c) or two-tailed paired Student's t-test (e). n.s., not significant.
Extended Data Fig. 4 Glioma xenograft calcium imaging with GCaMP6.
a, Confocal micrographs of xenografted SU-DIPG-XIII-FL cells expressing GCaMP6s-tdTomato into the hippocampus, and stained with DAPI (blue), HNA (green), and tdTomato nuclear tag (red). Merged image illustrates specificity of tdTomato tag to HNA+ cells. Scale bars, 20 µm. Immunostaining independently replicated in three mice. b, Spontaneous calcium transients in SU-DIPG-VI xenograft visualized by two-photon in situ calcium imaging (n = 5 mice). Representative frames shown. Red denotes glioma tdTomato nuclear tag; green denotes GCaMP6s. Scale bar, 50 µm. See also Supplementary Video 1. c, Trace of normalized GCaMP6s intensity over time in an individual xenografted glioma (SU-DIPG-VI) cell exhibiting an oscillatory spontaneous transient. Results were replicated across n = 3 mice. d, As in c, trace of normalized GCaMP6s intensity over time in an individual xenografted glioma (SU-DIPG-XIII-FL) cell exhibiting a less regular spontaneous transient. This type of transient is more frequently observed in glioma xenografts. Results were replicated across n = 3 mice. e, Individual xenografted glioma (SU-DIPG-XIII-FL) cellular responses to axonal stimulation before and after application of 0.5 µM TTX as measured by GCaMP6s intensity (n = 40 cells, 4 mice). Data are mean ± s.e.m. ****P < 0.0001, one-tailed Wilcoxon matched-pairs signed rank test.
Extended Data Fig. 5 Prolonged glioma currents and paediatric glioma tumour microtubes.
a, Left, time course of prolonged current block by 100 µM NASPM (red bar denotes duration of NASPM application; n = 8 cells, 5 mice). Data are mean ± s.e.m. Right, representative traces of evoked prolonged current (block) unaffected by NASPM (red). b, Quantification of data in a. Data are mean ± s.e.m; ns, not significant (two-tailed paired Student’s t-test). c, Alignment of phase-locked simultaneous recording of glioma prolonged potential with the field potential of firing neuronal population. d, Representative prolonged current traces with increasing stimulation intensity. Red denotes maximum intensity; blue and green denote intermediate intensities. Magnified view illustrates distinct spike-like waveforms consistent with responses to neuronal population firing. e, Relationship of extracellular field potential to magnitude of prolonged current (SU-DIPG-XIII-FL xenograft) illustrated by simultaneous field potential (fEPSP) and whole-cell glioma current-clamp recordings. f, Prolonged glioma potential amplitudes versus slope of fEPSPs elicited by electrical stimulation (10, 20, 30, 50, 70, 100 and 150 µA; R2 = 0.92; n = 14 cells or fields across 4 mice for each, except n = 11 cells or fields across 3 mice for 30 µA). g, Representative trace of potassium (K+)-induced prolonged current in SU-DIPG-XIII-FL xenografts (n = 9 cells, 2 mice). h, Effect of TBOA on prolonged current in glioma (SU-DIPG-XIII-FL) (n = 5 cells, 2 mice). (1) Representative trace of residual current left after application of d-AP5 and NBQX in prolonged response to stimulation. d-AP5 and NBQX are likely to reduce the prolonged current owing to the effect on CA1 pyramidal neurons, not through direct effect on the glioma cells themselves; (2) representative trace of residual current after trace 1 was then treated with glutamate transporter blocker 200 µM TBOA; (1 − 2) subtraction of trace 2 from trace 1 reveals a 2 pA current that can be accounted for by glutamate transporters. It should be noted that a small residual current remains. i, Synchronicity analysis of calcium peaks in glioma cells in Fig. 3 shown over the course of 10 min. Red lines indicate cells synchronized with one another at various time points during indicated period. j–o, Confocal micrographs of primary human glioma tissue samples illustrate density and length of nestin-immunopositive tumour microtubes. j–n, Primary human tissue samples of paediatric H3K27M+ DIPG, sampled at the time of autopsy. o, Analysis of primary tissue sample from an adult glioblastoma (SU-GBM092) illustrates the similarity of paediatric glioma to adult glioma. For all images, blue denotes DAPI; yellow denotes nestin; magenta denotes H3K27M (tumour-specific antigen). Scale bars, 20 µm (j–m, o), or 10 µm (n). Data independently replicated in three sections per sample.
Extended Data Fig. 6 Prolonged currents in paediatric glioma amplified by gap junction coupling.
a, Input resistance of non-responding and depolarizing cells (n = 29 fast EPSCs, n = 247 prolonged currents, n = 319 no response, data points above 1500 MΩ have been left off graph for clarity). b, Biocytin dye-filling illustrates coupling of xenografted glioma cells that exhibit prolonged currents (n = 7 slices, 2 mice). Red denotes streptavidin–biocytin; green denotes GFP. Scale bar, 100 µm. c, Prolonged currents in glioma markedly decreased by 100 µM CBX. Red box highlights magnified view of representative trace to illustrate residual slow current after application of CBX. d, Time course of prolonged current in glioma cell (black) and pyramidal neuron EPSC (blue) responses to the addition of CBX with subsequent washout of inhibitor (red bar denotes duration of CBX application; n = 6 glioma cells, 6 mice; n = 5 neurons, 2 mice). e, Quantification of the drop in current amplitude after addition of CBX (n = 19 cells, 11 mice; data also shown in Fig. 3g). f, Input resistance of cells in d in response to CBX. g, Quantification of data in f (n = 19 cells, 11 mice). h, Time course of prolonged current in glioma in response to addition of 100 µM meclofenamate (red bar denotes duration; n = 8 cells, 3 mice). i, Left, quantification of data in h; data also shown in Fig. 3g. Right, representative traces illustrating decrease in prolonged current amplitude after addition of meclofenamate (red). j, Input resistance of cells in h in response to addition of meclofenamate (n = 8 cells, 3 mice). k, Quantification of data in j. l, Representative frames from two-photon calcium imaging of in situ SU-DIPG-XIII-FL glioma xenografts illustrating spontaneous transients before (top) or after (bottom) addition of CBX. Red denotes tdTomato nuclear tag in glioma cells; green denotes GCaMP6s. Scale bar, 50 µm. Results replicated across three mice. m, Synchronicity analyses of spontaneous calcium transients before (top) or after (bottom) addition of CBX. n, Representative trace of GCaMP6s peak intensity over time in a single cell before (black) and after (red) addition of CBX (n = 3 mice). o, Synchronicity scores of individual cells within glioma xenografts before and after addition of CBX (n = 164 cells, 3 mice). y axis represents synchronicity score; x axis represents number of cells with specific score. Data are mean ± s.e.m. (a, d–k). **P < 0.01, ****P < 0.0001, one-way ANOVA with Tukey’s post hoc analysis (a) or one-tailed Wilcoxon matched-pairs signed rank test (e, g, i, k, o).
Extended Data Fig. 7 Heterogeneity of glioma cell electrophysiological response and validation of ChR2 and GluA2-DN function in glioma.
a, Electrophysiological responses by patient-derived pediatric high-grade glioma (pHGG) model. Number of whole-cell patch-clamp recordings from glioma cells in xenografted hippocampal slices categorized by electrophysiological response to axonal electrical stimulation. b, Demonstration of depolarizing inward current in SU-DIPG-XIII-FL-ChR2 cells in response to single stimulation and 20 Hz pulses of blue light as measured in current clamp (top) and voltage clamp (bottom). c, Proliferation index of xenografted SU-DIPG-XIII-FL-YFP control glioma cells (no opsin expressed) in response to blue-light or mock stimulation as measured by the proportion of GFP+/HNA+ cells expressing Ki67 24 h after five optogenetic stimulation sessions (n = 3 mice, mock stim; n = 4 mice, stim). d, Quantification of cleaved caspase-3 in xenografted SU-DIPG-XIII-FL-YFP control glioma cells in response to blue-light or mock stimulation as measured by total number of HNA+ cells co-labelled with cleaved caspase-3 (n = 3 mice per group). e, As in d, but for xenografted SU-DIPG-XIII-FL-ChR2 glioma cells (n = 3 mice, mock stim; n = 4 mice, stim). f, g, Validation of GluA2-DN-expressing construct. Representative traces of whole-cell voltage-clamp recording of wild-type (black) and GluA2-DN-expressing (grey) SU-DIPG-VI (f) or SU-DIPG-XIII-FL (g) cells in response to 500µM (S)-AMPA (n = 6 cells for each). SU-DIPG-XIII-FL cells are unable to homogeneously express the dominant construct, and therefore may be gap junction-coupled to wild-type GluA2-expressing cells, which may account for the remaining current in the illustrated trace (g). Incorporation of the GluA2-DN construct thus results in abrogated AMPAR-dependent depolarization. Data are mean ± s.e.m. (c–e). NS, not significant (two-tailed unpaired Student’s t-test).
Extended Data Fig. 8 Glioma AMPAR function in vitro, in co-culture and in vivo.
a, Kaplan–Meier survival curves of a second cohort of mice orthotopically xenografted with control GFP-only or GluA2-DN-GFP-overexpressing cells (SU-DIPG-XIII-P∗ xenograft model; n = 5 mice per group). b, Representative serial coronal sections of mouse brains bearing SU-DIPG-XIII-FL xenografts expressing either control GFP construct (left) or GluA2-DN-GFP construct (right). Grey denotes MBP; white denotes glioma-GFP. c, Proliferation indices of SU-DIPG-XIII-FL cells at baseline in neuronal medium, in response to 10 µM NBQX, in co-culture with neurons, or in co-culture with neurons in the presence of 10 µM NBQX (n = 3 biological replicates per group, except n = 4 for baseline). d, Representative images of neuron–glioma co-cultures in the presence and absence of NBQX. Green denotes neurofilament (neuronal processes); red denotes nestin (glioma cell processes); white denotes Ki67. Scale bar, 50 µm. e, In vitro growth analysis of glioma cells expressing control GFP or GluA2-DN-GFP monitored over 3 days. f, In vitro apoptosis analysis of glioma cells expressing control GFP or GluA2-DN-GFP as measured by percentage of total cells co-stained with cleaved caspase-3. g, 3D Matrigel invasion assay in control GFP-expressing and GluA2-DN-GFP-expressing SU-DIPG-XIII-FL cells 72 h after seeding. h, Representative images of data in g at time 0 h (left) and 72 h (right) in control GFP-expressing (top) and GluA2-DN-GFP-expressing cells (bottom). Scale bars, 1,000 µm. i, 3D migration in control GFP-expressing and GluA2-DN-GFP-expressing SU-DIPG-XIII-FL cells 72 h after seeding. j, Representative images of data in i at time 0 h (left) and 72 h (right) in control GFP-expressing (top) and GluA2-DN-GFP-expressing cells (bottom). Scale bars, 1,000 µm. k, Representative confocal micrographs illustrating proliferating SU-DIPG-VI cells in vehicle or perampanel-treated mice (n = 8 mice per group). Red denotes human nuclei (HNA); white denotes Ki67. Scale bar, 50 µm. l, IVIS bioluminescence analysis of overall tumour growth in SU-DIPG-XIII-FL xenografts treated with vehicle or meclofenamate over a two-week period (n = 5 mice per group). Data represented as fold change in total flux. Data shown are mean ± s.e.m. (c, e–g, i, l). For analyses in d–j, n = 3 biological replicates. *P < 0.05, **P < 0.01, ****P < 0.0001, two-tailed log rank analyses (a), one-way ANOVA with Tukey’s post hoc analysis (c), or two-tailed unpaired Student’s t-test (f, g, i, l).
Extended Data Fig. 9 Hyperexcitability in the glioma microenvironment and working model of neuron–glioma interactions in the tumour microenvironment.
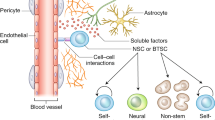
a, Individual channel electrocorticography signals (mean high-gamma frequency-filtered power) in each of healthy-appearing, tumour core, and tumour-infiltrated brain (n = 23, 29 and 51 total channels, respectively) across three human subjects, as in Fig. 5a, b. Data shown are mean ± s.e.m. ****P < 0.0001, one-way ANOVA with Tukey’s post hoc analysis. b, Working model of glioma integration into neural circuity, with hyperexcitability of neurons (grey) exacerbating activity-dependent mechanisms of glioma (green) growth. (1) neuron-to-glioma synapses (synaptic vesicles in red; AMPARs in grey); (2) inward potassium (K+) current (blue; potassium channel in grey); (3) gap junction (white) coupling in glioma amplifies current.
Supplementary information
Supplementary Information
This file contains Additional Statistical Information.
Supplementary Figures
This file contains original western blot scans.
Supplementary Video 1:
In situ two-photon imaging of glioma expressing genetically encoded calcium indicator. Live in situ two-photon imaging of H3K27M+ DMG xenografts expressing the genetically encoded calcium indicator GCaMP6s (SU-DIPGXIII-FL-GCaMP6s cells) xenografted into the CA1 region of the mouse hippocampus. Glioma nuclei are labelled red with td-Tomato; spontaneous calcium transients visualized in green. Data were independently replicated in 14 mice. Video is taken at ~1.5 Hz time compressed 15x (30fps).
Supplementary Video 2:
In situ two-photon imaging of glioma expressing genetically encoded calcium indicator in response to electrical stimulation of local afferents. Live in situ two-photon imaging of H3K27M+ DMG xenografts expressing the genetically encoded calcium indicator GCaMP6s (SU-DIPGXIII-FL-GCaMP6s cells) xenografted into the CA1 region of the mouse hippocampus in response to local Schaffer collateral afferent stimulation. Glioma nuclei are labelled red with td-Tomato; calcium transients visualized in green. Data were independently replicated in 4 mice. Video is taken at ~1.5 Hz time compressed 15x (30fps). Time of stimulation is annotated “stim”.
Supplementary Video 3:
In situ two-photon imaging of glioma expressing genetically encoded calcium indicator after addition of gap junction blocker carbenoxolone. Live in situ two-photon imaging of H3K27M+ DMG xenografts expressing the genetically encoded calcium indicator GCaMP6s (SU-DIPGXIII-FL-GCaMP6s cells) xenografted into the CA1 region of the mouse hippocampus. Spontaneous calcium transients shown before and after addition of gap junction blocker carbenoxolone (100µM). Glioma nuclei are labelled red with td-Tomato; calcium transients visualized in green. Data were independently replicated in 4 mice. Video is taken at ~1.5 Hz time compressed 15x (30fps). Addition of inhibitor annotated as “+CBX.”.
Rights and permissions
About this article
Cite this article
Venkatesh, H.S., Morishita, W., Geraghty, A.C. et al. Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019). https://doi.org/10.1038/s41586-019-1563-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1563-y
This article is cited by
-
PerSurge (NOA-30) phase II trial of perampanel treatment around surgery in patients with progressive glioblastoma
BMC Cancer (2024)
-
A phase Ib/II randomized, open-label drug repurposing trial of glutamate signaling inhibitors in combination with chemoradiotherapy in patients with newly diagnosed glioblastoma: the GLUGLIO trial protocol
BMC Cancer (2024)
-
Combining a noble gas with radiotherapy: glutamate receptor antagonist xenon may act as a radiosensitizer in glioblastoma
Radiation Oncology (2024)
-
Gliomas: a reflection of temporal gliogenic principles
Communications Biology (2024)
-
Pan-cancer ion transport signature reveals functional regulators of glioblastoma aggression
The EMBO Journal (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.