Abstract
The family of bacterial SidE enzymes catalyses phosphoribosyl-linked serine ubiquitination and promotes infectivity of Legionella pneumophila, a pathogenic bacteria that causes Legionnaires’ disease1,2,3. SidE enzymes share the genetic locus with the Legionella effector SidJ that spatiotemporally opposes the toxicity of these enzymes in yeast and mammalian cells, through a mechanism that is currently unknown4,5,6. Deletion of SidJ leads to a substantial defect in the growth of Legionella in both its natural hosts (amoebae) and in mouse macrophages4,5. Here we demonstrate that SidJ is a glutamylase that modifies the catalytic glutamate in the mono-ADP ribosyl transferase domain of the SdeA, thus blocking the ubiquitin ligase activity of SdeA. The glutamylation activity of SidJ requires interaction with the eukaryotic-specific co-factor calmodulin, and can be regulated by intracellular changes in Ca2+ concentrations. The cryo-electron microscopy structure of SidJ in complex with human apo-calmodulin revealed the architecture of this heterodimeric glutamylase. We show that, in cells infected with L. pneumophila, SidJ mediates the glutamylation of SidE enzymes on the surface of vacuoles that contain Legionella. We used quantitative proteomics to uncover multiple host proteins as putative targets of SidJ-mediated glutamylation. Our study reveals the mechanism by which SidE ligases are inhibited by a SidJ–calmodulin glutamylase, and opens avenues for exploring an understudied protein modification (glutamylation) in eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Mass spectrometry data are available from the Proteomics Identification (PRIDE) database (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier29 PXD014362. Cryo-EM structure coordinates are available from the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB) under accession codes 6S5T and EMD-10100, respectively. Full gel source data can be found in Supplementary Fig. 1. The data that support the findings of this study are available from the corresponding authors upon request.
References
Hubber, A. & Roy, C. R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 (2010).
Qiu, J. et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 (2016).
Bhogaraju, S. et al. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649.e13 (2016).
Jeong, K. C., Sexton, J. A. & Vogel, J. P. Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog. 11, e1004695 (2015).
Liu, Y. & Luo, Z.-Q. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75, 592–603 (2007).
Havey, J. C. & Roy, C. R. Toxicity and SidJ-mediated suppression of toxicity require distinct regions in the SidE family of Legionella pneumophila effectors. Infect. Immun. 83, 3506–3514 (2015).
Urbanus, M. L. et al. Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12, 893 (2016).
Qiu, J. et al. A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res. 27, 865–881 (2017).
Bähler, M. & Rhoads, A. Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107–113 (2002).
Solntsev, S. K., Shortreed, M. R., Frey, B. L. & Smith, L. M. Enhanced global post-translational modification discovery with MetaMorpheus. J. Proteome Res. 17, 1844–1851 (2018).
Black, M. H. et al. Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364, 787–792 (2019).
Starovasnik, M. A., Davis, T. N. & Klevit, R. E. Similarities and differences between yeast and vertebrate calmodulin: an examination of the calcium-binding and structural properties of calmodulin from the yeast Saccharomyces cerevisiae. Biochemistry 32, 3261–3270 (1993).
Ishida, M. et al. The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property. Biochemistry 41, 15536–15542 (2002).
Bardill, J. P., Miller, J. L. & Vogel, J. P. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56, 90–103 (2005).
Drum, C. L. et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 415, 396–402 (2002).
Wolff, J., Cook, G. H., Goldhammer, A. R. & Berkowitz, S. A. Calmodulin activates prokaryotic adenylate cyclase. Proc. Natl Acad. Sci. USA 77, 3841–3844 (1980).
Kalayil, S. et al. Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557, 734–738 (2018).
Grumati, P. et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6, e25555 (2017).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Grynkiewicz, G., Poenie, M. & Tsien, R. Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 (1985).
Tegunov, D. & Cramer, P. Real-time cryo-EM data pre-processing with Warp. Preprint at https://www.biorxiv.org/content/10.1101/338558v1 (2018).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Burnley, T., Palmer, C. M. & Winn, M. Recent developments in the CCP-EM software suite. Acta Crystallogr. D 73, 469–477 (2017).
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Acknowledgements
We thank J. Vogel for the ΔsidJ Legionella strain; Z.-Q. Luo for SidJ proteins purified from Legionella, and for providing ΔsidE and wild-type Legionella strains; S. Rodriguez and S. Gharbi for technical assistance; Y. Liu for help with deubiquitination assays; D. Höller, H. Marei and K. Koch for critical comments on the manuscript; and S. Knapp and V. Doetsch for discussion and advice. This work was supported by the DFG-funded Collaborative Research Centre on Selective Autophagy (SFB 1177), by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 742720), by the DFG-funded Cluster of Excellence ‘Macromolecular Complexes’ (EXC115) and by the DFG-funded SPP 1580 program ‘Intracellular Compartments as Places of Pathogen-Host-Interactions’ (to I.D.).
Author information
Authors and Affiliations
Contributions
S.B. and I.D. conceived and supervised the project. S.B. performed mammalian SidJ and SdeA co-expression and ε-NAD+ hydrolysis assays. F.B. performed all the mass spectrometry experiments with the help of V.M. and S.B. in sample preparation. R.M. performed Legionella infection experiments and prepared samples for mass spectrometry. M.A. performed in vitro ε-NAD+ hydrolysis assays, isothermal titration calorimetry and protein purifications for biochemistry and cryo-EM. M.M.P. and W.P.G. prepared cryo-EM grids, collected and processed data and performed the model refinement. J.L.-M. performed yeast experiments. S.K. performed protein purifications. D.S. performed protein purifications and in vitro SidJ–CaM pulldown experiments. S.B. and I.D. analysed the data and wrote the manuscript with input from all the co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Friedrich Förster, Elizabeth Hartland, Carsten Janke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
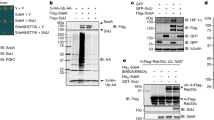
Extended Data Fig. 1 SidJ does not possess intrinsic deubiquitinase activity.
a, Genetic locus of sdeC-orf2-sidj-sdeB-sdeA in the Legionella genome. b, Left, GFP–SidJ that was ectopically expressed and purified from HEK293T cells was incubated with canonical HA–ubiquitin chains purified from mammalian cells. The canonical deubiquitinase USP2 was used as a positive control. Right, GFP or GFP–SidJ was incubated with purified SdeA–ubiquitinated Rab33b. The experiment was repeated twice independently, with similar results. c, Full-length SidJ was incubated with various substrates modified with canonical ubiquitination or phosphoribosyl-linked ubiquitination, to probe the cleavage activity. USP2 was used as a positive control for cleaving canonical ubiquitin chains. The experiment was repeated twice independently, with similar results. d, SidJ purified from Legionella was analysed by mass spectrometry, and protein quantification was performed using the MaxQuant iBAQ algorithm. The experiment was repeated twice independently, with similar results.
Extended Data Fig. 2 SidJ binds to CaM.
a, GFP and GFP–SidJ were ectopically expressed in HEK293T cells, and immunoprecipitated. The samples were analysed by SDS–PAGE, followed by Coomassie staining. The experiment was repeated twice independently, with similar results. b, Purified SdeA from HEK293T cells that express SdeA alone, or in combination with SidJ or SidJ(ΔIQ) (here labelled SidJ(1–819)), was used in ε-NAD+ hydrolysis assays. The experiment was repeated twice independently, with similar results. c, A549 cells infected with wild-type L. pneumophila for different time periods were loaded with Fura2AM for 30 min at 37 °C, followed by ratiometric measurement of intracellular Ca2+ using a plate reader. Infection with bacteria did not change total Ca2+ levels in the cell. n = 3 biologically independent experiments, Data points indicate mean and error bars represent s.d. d, A549 cells expressing endoplasmic reticulum–GFP and endoplasmic reticulum–cepia were infected with L. pneumophila (Ds-Red lp02) followed by time-lapse imaging. Fluorescence intensity of endoplasmic reticulum–cepia at each region is proportional to local Ca2+ levels. Endoplasmic reticulum–GFP fluorescence is independent of Ca2+ concentration. The endoplasmic reticulum has a heterogenous and dynamic distribution of Ca2+. The bacteria make transient contacts with the endoplasmic reticulum, and may be influenced by local Ca2+ fluxes in the cell. Endoplasmic reticulum–cepia is marked in red, endoplasmic reticulum–GFP in green and bacteria are marked by white dotted lines. Time-lapse images were taken at 1-s intervals for 2 min. Images shown in the montage are at 10-s intervals. The experiment was repeated three times independently, with similar results. e, Crystal structure of SdeA (PDB 5YIM) is shown in cartoon representation, highlighting the missing peptide of SdeA in SidJ-treated samples (shown in red). The solvent-exposed part of this peptide that contains the catalytic glutamate (E860, shown in green) is marked.
Extended Data Fig. 3 Mass spectra showing glutamylation of E860 of SdeA.
a, Annotated mass spectra for di-glutamylation of E860 of SdeA from samples of immunoprecipitated GFP–SdeA co-expressed with SidJ. b, In vitro glutamylation of SdeA was carried in various conditions (as indicated), followed by ε-NAD+ hydrolysis assays to measure the ubiquitin–ADP ribosylation activity of SdeA. The experiment was repeated three times independently, with similar results.
Extended Data Fig. 4 In vitro glutamylation of SdeA.
a, Samples of in vitro glutamylation reactions that contained SdeA and SidJ, with or without ATP, were TMT-labelled and analysed by quantitative mass spectrometry. Mono- and di-glutamylation of the catalytic E860 of SdeA was enriched in samples that contained ATP. In vitro glutamylation was performed in n = 3 biologically independent experiments. Samples were labelled with TMT six-plex reagent and analysed in one liquid chromatography–mass spectrometry run. Significant differences between samples were detected by a two-sided Student’s t-test. b, Annotated mass spectra for mono-glutamylation of E860 of SdeA, from SidJ glutamylase in vitro reactions. c, Annotated mass spectra for di-glutamylation of E860 of SdeA, from SidJ glutamylase in vitro reactions.
Extended Data Fig. 5 Analysis of isobaric glutamylated peptide species.
a, Extracted ion chromatogram of +4-charged QVGRHGEGTESEFSVYLPEDVALVPVK peptide with +1 glutamate, showing that there are no other co-existing mono-glutamylated versions of the catalytic peptide (besides glutamylation of E860). b, Extracted ion chromatogram of the catalytic peptide plus two glutamates (charge +4) is separated into three peaks that could be assigned to di-glutamylation of E860, as well as two mono-glutamylations on the peptide on E860 and E857, and on E860 and E862. Annotated spectra are shown below. c, Extracted ion chromatogram of the catalytic peptide plus three glutamates (charge +4) is separated into three different peaks that could be assigned to di-glutamylation of E860, plus mono-glutamylation of E857 and a parallel mono-glutamylation of E857, E860 and E862; a third peak could not be clearly assigned. Annotated spectra of the annotated species are shown below. In a–c, in vitro glutamylation and label-free liquid chromatography–mass spectrometry analysis were performed in three biologically independent experiments with similar results. Corresponding quantitative information is shown in Fig. 3d.
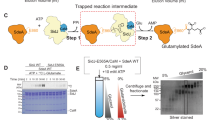
Extended Data Fig. 6 Cryo-EM data processing and 3D reconstruction.
a, Size-exclusion profile of SidJ–CaM complex. Elution fractions were analysed by SDS–PAGE. Marked fractions were used for cryo-EM sample preparation. This experiment was repeated three times independently, with similar results. b, A representative electron micrograph for the cryo-EM dataset collected. c, Reference-free representative 2D class averages of the SidJ–CaM complex. Secondary structure features are visible in projection images. The number (n) of particles used to obtain a 2D class average is mentioned in each subpanel. d, Gold-standard FSC30 plot between two independently refined half-maps, FSC0.143 = 4.15-Å resolution. As expected, FSC between phase-randomized half-maps show a rapid drop of correlation beyond randomization point. e, Crystal structure of SidJ–CaM (PDB 6OQQ) is fitted into the cryo-EM 3D reconstruction. f, Part of d, magnified to highlight the difference between the crystal structure and the cryo-EM map.
Extended Data Fig. 7 Cryo-EM single-particle analysis pipeline.
Data-processing strategy for cryo-EM of the SidJ–CaM complex. Particle-picking on 2,423 micrographs (using WARP) resulted in the identification of 1,500,000 particles. The particle coordinates were imported into Relion and particles were extracted with a twofold binning factor. After 2D classification and an initial 3D classification, a 3D class with clear secondary structure features and 370,000 particles was identified. The particles of this class were re-extracted with full pixel size and 3D-refined, resulting in a 4.52-Å model. Two additional rounds of 3D classification and 3D refinement improved the resolution of the model to 4.15 Å. Final particle polishing and CTF refinement of the remaining particles did not result in a nominal improvement of the resolution.
Extended Data Fig. 8 Cryo-EM model refinement.
a, FSC between model and the map and cross-validation of the model fitting. FSC0.5 = 4.3 Å for the model versus map (sum). Half-map cross validation procedure31 does not show overfitting in the refined model. b, Local resolution analysis (using Relion) shows variation in the map resolution ranging from 3.9 to 5.1 Å. c, Overview of the model fitting into the map in the same orientation as in b. d, An example of the cryo-EM map quality with the atomic model fitted in, showing clear density for side chains.
Extended Data Fig. 9 Ligand-binding sites of SidJ–CaM complex.
a, Crystal structure of complex between SidJ and yeast CaM (PDB 6OQQ) is shown, marking the two proposed catalytic sites and the bound ligands. b, Electron-microscopy map showing cryo-EM density in the migrated pocket fitted with AMPPNP. c, Electron-microscopy map showing unassigned cryo-EM density in the canonical pocket.
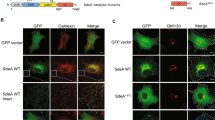
Extended Data Fig. 10 SidJ-dependent glutamylation during Legionella infection.
a, Raw264.7 macrophages were infected with wild-type, ΔSidJ or ΔSidE Legionella for 3 h. Lysates were used for immunoprecipitation with polyglutamylation antibody, followed by immunoblotting with SdeA. ni, samples that were not infected with bacteria. This experiment was repeated twice independently, with similar results. b, A549 cells were infected with different strains of L. pneumophila for 3 h. Cells were fixed and immunostained with antibodies against calnexin and polyglutamylation (GT335). DAPI staining marks the nucleus and cytosolic bacteria. Yellow arrows indicate bacteria in infected cells. The ROI is defined as calnexin-stained LCV. ROIs of 80 × 100 μm2 were chosen in the perinuclear region of cells, followed by quantification of Mander’s coefficient (m) using the Coloc2 plugin in FIJI. m represents the fraction of calnexin-positive LCVs that are also positive for polyglutamylation. Centre lines show the medians; box limits indicate the 25th and 75th percentiles (as determined by R software); whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles; and outliers are represented by dots. Number of ROIs (n) = 80 from 30 cells was used for quantification. ***P < 0.001 by two-tailed type-3 Student’s t-test. P value (wild type versus ΔsidJ) = 6.18 × 10−29; P value (wild type versus ΔsidE) = 1.09 × 10−5. This experiment was repeated twice independently, with similar results. c, Glutamylated proteins were isolated from wild-type, ΔsidE and ΔsidJ Legionella infection experiments using GT335 antibody and quantified using mass spectrometry. Correlation between wild-type versus ΔsidJ and ΔsidE versus ΔsidJ quantifications are plotted (inset), showing the most-correlated proteins in these two quantifications. Legionella infection and label-free liquid chromatograph–mass spectrometry analysis was performed with n = 3 biologically independent experiments. Significant differences between samples were detected by a corrected, two-sided Student's t-test with permutation-based false-discovery rate of 0.05. Proteins were labelled as significant if they were above the false-discovery-rate threshold of 0.05 in at least one comparison (ΔsidE and wild-type Legionella compared to ΔsidJ-infected cells). Proteins with a log2 ratio above two (mean) in wild-type samples were labelled as highly enriched compared to ΔsidJ-infected cells in samples from wild-type and ΔsidE-infected cells.
Supplementary information
Supplementary Information
A description of the ligand binding sites in the Cryo-EM structure of SidJ/CaM, Supplementary Table 1 and Supplementary Figure 1, the uncropped gels.
Video 1: Calcium dynamics at the legionella-ER contact sites during infection
A549 cells expressing ER-GFP and ER-cepia were infected with Legionella pneumophilia (Ds-Red lp02) followed by time-lapse imaging. Fluorescence intensity of ER-cepia at each region is proportional to local Ca2+ levels. This video corresponds to the time-lapse figure presented in Extended Data Fig. 2d.
Rights and permissions
About this article
Cite this article
Bhogaraju, S., Bonn, F., Mukherjee, R. et al. Inhibition of bacterial ubiquitin ligases by SidJ–calmodulin catalysed glutamylation. Nature 572, 382–386 (2019). https://doi.org/10.1038/s41586-019-1440-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1440-8
This article is cited by
-
Legionella metaeffector MavL reverses ubiquitin ADP-ribosylation via a conserved arginine-specific macrodomain
Nature Communications (2024)
-
Structural basis for the toxicity of Legionella pneumophila effector SidH
Nature Communications (2023)
-
An expanded lexicon for the ubiquitin code
Nature Reviews Molecular Cell Biology (2023)
-
Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis
Nature Structural & Molecular Biology (2023)
-
Polyglutamylation: biology and analysis
Amino Acids (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.