Abstract
Almost all eukaryote life forms have now been placed within one of five to eight supra-kingdom-level groups using molecular phylogenetics1,2,3,4. The ‘phylum’ Hemimastigophora is probably the most distinctive morphologically defined lineage that still awaits such a phylogenetic assignment. First observed in the nineteenth century, hemimastigotes are free-living predatory protists with two rows of flagella and a unique cell architecture5,6,7; to our knowledge, no molecular sequence data or cultures are currently available for this group. Here we report phylogenomic analyses based on high-coverage, cultivation-independent transcriptomics that place Hemimastigophora outside of all established eukaryote supergroups. They instead comprise an independent supra-kingdom-level lineage that most likely forms a sister clade to the ‘Diaphoretickes’ half of eukaryote diversity (that is, the ‘stramenopiles, alveolates and Rhizaria’ supergroup (Sar), Archaeplastida and Cryptista, as well as other major groups). The previous ranking of Hemimastigophora as a phylum understates the evolutionary distinctiveness of this group, which has considerable importance for investigations into the deep-level evolutionary history of eukaryotic life—ranging from understanding the origins of fundamental cell systems to placing the root of the tree. We have also established the first culture of a hemimastigote (Hemimastix kukwesjijk sp. nov.), which will facilitate future genomic and cell-biological investigations into eukaryote evolution and the last eukaryotic common ancestor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw reads of Spironema and Hemimastix transcriptomes are deposited in GenBank under accession codes SRR6032743 and SRR6032744, respectively. The assembled Hemimastix and Spironema transcriptomes, 351 individual-gene alignments (104 taxa), concatenated and trimmed alignments and tree-files for the 104-taxon, 61-taxon, 58-nLB, 58-nDP, 61-SR4 and 61-SFSR datasets, alignments and tree files for non-universal ancient genes, raw light microscopy and scanning electron microscopy images, and the SSU rDNA alignment and tree-files have been deposited in Dryad (https://doi.org/10.5061/dryad.n5g39d7). The partial SSU rDNA gene sequence of H. kukwesjijk strain BW2H is deposited in GenBank, under accession code MF682191. This publication has been registered with the ZooBank database (http://zoobank.org/) with the Life Science Identifier urn:lsid:zoobank.org:pub:4BA2A83C-8363-4EBE-A9C7-097CA470F9FB, and the name Hemimastix kukwesjijk has been deposited in Zoobank with the Life Science Identifier urn:lsid:zoobank.org:act:32E12332-A418-40E2-BF4C-F2BFD94BF4CF.
References
Burki, F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 6, a016147 (2014).
Worden, A. Z. et al. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (2015).
Burki, F. et al. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. R. Soc. Lond. B 283, 20152802 (2016).
Simpson, A. G. B. & Eglit, Y. in Encyclopedia of Evolutionary Biology Vol. 3 (ed. Kliman, R. M.) 344–360 (Elsevier, Amsterdam, 2016).
Klebs, G. Flagellatenstudien (Akademische Verlags-Gesellschaft, Leipzig, 1893).
Foissner, W., Blatterer, H. & Foissner, I. The Hemimastigophora (Hemimastix amphikineta nov. gen., nov. spec.), a new protistan phylum from Gondwanian soils. Eur. J. Protistol. 23, 361–383 (1988).
Foissner, I. & Foissner, W. Revision of the family Spironemidae Doflein (Protista, Hemimastigophora), with description of two new species, Spironema terricola n. sp. and Stereonema geiseri n. g., n. sp. J. Eukaryot. Microbiol. 40, 422–438 (1993).
Yubuki, N. et al. Morphological identities of two different marine stramenopile environmental sequence clades: Bicosoeca kenaiensis (Hilliard, 1971) and Cantina marsupialis (Larsen and Patterson, 1990) gen. nov., comb. nov. J. Eukaryot. Microbiol. 62, 532–542 (2015).
Brown, M. W. et al. Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proc. R. Soc. Lond. B 280, 20131755 (2013).
Zhao, S. et al. Collodictyon—an ancient lineage in the tree of eukaryotes. Mol. Biol. Evol. 29, 1557–1568 (2012).
Cavalier-Smith, T. et al. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol. Phylogenet. Evol. 81, 71–85 (2014).
Cavalier-Smith, T. A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 73, 203–266 (1998).
Cavalier-Smith, T. in The Flagellates, The Systematics Association Special Volume Series 59 (eds Leadbeater, B. S. C. & Green, J. C.) 361–390 (Taylor & Francis, London, 2000).
Cavalier-Smith, T., Lewis, R., Chao, E. E., Oates, B. & Bass, D. Morphology and phylogeny of Sainouron acronematica sp. n. and the ultrastructural unity of Cercozoa. Protist 159, 591–620 (2008).
Speijer, D., Lukeš, J. & Eliáš, M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl Acad. Sci. USA 112, 8827–8834 (2015).
de Mendoza, A. et al. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl Acad. Sci. USA 110, E4858–E4866 (2013).
Fukasawa, Y., Oda, T., Tomii, K. & Imai, K. Origin and evolutionary alteration of the mitochondrial import system in eukaryotic lineages. Mol. Biol. Evol. 34, 1574–1586 (2017).
Sebé-Pedrós, A., Grau-Bové, X., Richards, T. A. & Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 6, 290–305 (2014).
Barlow, L. D., Nývltová, E., Aguilar, M., Tachezy, J. & Dacks, J. B. A sophisticated, differentiated Golgi in the ancestor of eukaryotes. BMC Biol. 16, 27 (2018).
He, D. et al. An alternative root for the eukaryote tree of life. Curr. Biol. 24, 465–470 (2014).
Katz, L. A., Grant, J. R., Parfrey, L. W. & Burleigh, J. G. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst. Biol. 61, 653–660 (2012).
Derelle, R. & Lang, B. F. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29, 1277–1289 (2012).
Derelle, R. et al. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl Acad. Sci. USA 112, E693–E699 (2015).
Richards, T. A. & Cavalier-Smith, T. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118 (2005).
Kolisko, M., Boscaro, V., Burki, F., Lynn, D. H. & Keeling, P. J. Single-cell transcriptomics for microbial eukaryotes. Curr. Biol. 24, R1081–R1082 (2014).
Yoon, H. S. et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717 (2011).
Gawryluk, R. M. R. et al. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 26, 3053–3059 (2016).
Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014).
Caron, D. A. et al. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 15, 6–20 (2017).
Krabberød, A. K. et al. Single cell transcriptomics, mega-phylogeny, and the genetic basis of morphological innovations in Rhizaria. Mol. Biol. Evol. 34, 1557–1573 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Huse, S. M. et al. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics 15, 41 (2014).
de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
BioMarKs Consortium. BioMarKs data portal http://www.biomarks.eu (2011).
Mahé, F. et al. Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat. Ecol. Evol. 1, 0091 (2017).
Marquardt, M., Vader, A., Stübner, E. I., Reigstad, M. & Gabrielsen, T. M. Strong seasonality of marine microbial eukaryotes in a high-arctic fjord (Isfjorden, in West Spitsbergen, Norway). Appl. Environ. Microbiol. 82, 1868–1880 (2016).
Geisen, S. et al. Metatranscriptomic census of active protists in soils. ISME J. 9, 2178–2190 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Berger, S. A. & Stamatakis, A. Aligning short reads to reference alignments and trees. Bioinformatics 27, 2068–2075 (2011).
Matsen, F. A., Kodner, R. B. & Armbrust, E. V. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 538 (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Brown, M. W. et al. Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biol. Evol. 10, 427–433 (2018).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Wang, H. C., Minh, B. Q., Susko, E. & Roger, A. J. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216–235 (2018).
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009).
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004).
Susko, E. & Roger, A. J. On reduced amino acid alphabets for phylogenetic inference. Mol. Biol. Evol. 24, 2139–2150 (2007).
Brown, J. W., Walker, J. F. & Smith, S. A. Phyx: phylogenetic tools for unix. Bioinformatics 33, 1886–1888 (2017).
Eddy, S. R. Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Foissner, W. & Foissner, I. in An Illustrated Guide to the Protozoa 2nd edn (eds Lee, J. J. et al.) 1185–1186 (Society of Protozoologists and Allen Press, Lawrence, 2002).
Zolffel, M. & Skibbe, O. Rediscovery of the multiflagellated protist Paramastix conifera Skuja 1948 (Protista incertae sedis). Nova Hedwigia 65, 443–452 (1997).
Acknowledgements
The authors thank P. Li and P. Scallion (Dalhousie University) for assistance with electron microscopy, M. Dlutek (Dalhousie University) for Illumina sequencing, S. Geisen (Wageningen University) for providing parsed metatranscriptomic data, F. Mahé (CIRAD, Montpellier) for access to and parsing much of the V4 data, M. Brown (Mississippi State) for the seed phylogenomic dataset, A. Sebé-Pedrós (Weizmann Institute of Science) for the seed myosin alignments, M. Kolisko (Institute of Parasitology, Czech Academy of Sciences) for data handling scripts, B. Q. Minh (University of Vienna) for substantial help with phylogenomic analyses and troubleshooting in IQ-TREE, and R. Lewis (Nova Scotia Museum) and B. Francis for advice on Mi’kmaq tradition and language. This work was supported by CIFAR, NSERC grant 298366-2014 to A.G.B.S. and NSERC grant 2016-016792 to A.J.R.
Reviewer information
Nature thanks I. Ruiz-Trillo and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Y.E. isolated the organisms and cultivated H. kukwesjijk. Y.E. and G.L. undertook the microscopy. G.L. performed the single-cell transcriptomics. Y.E., G.L. and E.M.B. analysed the rDNA and environmental sequence data. G.L., L.E., Y.E. and A.G.B.S. assembled the phylogenomic datasets. G.L., L.E. and A.J.R. performed phylogenomic analyses. L.E. and Y.E. performed the gene presence analyses. G.L., Y.E. and A.G.B.S. wrote the manuscript, with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
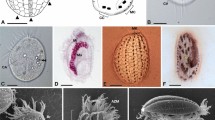
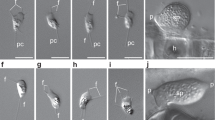
Extended Data Fig. 1 Light micrographs of studied hemimastigotes.
a–m, Spironema cf. multiciliatum (a) and Hemimastix kukwesjijk (b–m) differential interference contrast micrographs of live cells. a, Two views of a Spironema cf. multiciliatum cell, with inset that details the posterior end. Note the nucleus (marked by ‘n’), the detail of one of the posterior flagella (marked by an arrow, in the inset) and small contractile vacuole (cv, in inset), as well as posterior tail (line in inset). b, c, Optical sections through one H. kukwesjijk cell, detailing the notches from which flagella emerge (arrowheads), a section through the capitulum (marked with a ‘c’) and a conspicuous contractile vacuole in the cell posterior (shown in b). d, Surface view of one of the two thecal plates. e–g, Optical cross-sections of different cells showing the capitulum (e), mid-body region with rotationally symmetrical plate overlap (f) and the posterior (g) with radial arrangement of the posterior-most flagella. h–j, Pseudoseries that illustrates the feeding process, showing the progression of prey-ingestion stages. Note the widening capitulum and beginning of formation of the phagocytic vacuole. k, Same cell as in j, showing the anterior flagella curving forward to surround prey (seen especially in early feeding). l, m, Dividing cells, showing the diagonal symmetry of short new rows (nr) and longer old rows (or) of flagella, as well as the daughter nuclei (n). Scale bar, 10 μm.
Extended Data Fig. 2 Scanning electron microscopy images of H. kukwesjijk.
a, Feeding cell, general view (anterior to left; note the prey item attached to capitulum). b, Close-up of anterior end showing ingestion in progress at the capitulum. c, Discharged extrusomes (ex; triggered by the fixation process) along margin of the capitulum (compare to undischarged extrusomes in Fig. 1d). d, Dividing cells, with the left-most cell clearly showing the old row of full-length flagella (or) and the new row with short flagella (nr). Scale bars, 5 μm (a, d), 2 μm (b, c).
Extended Data Fig. 3 SSU rDNA phylogeny of eukaryotes.
Phylogeny inferred from 111 taxa and 1,252 sites under the GTR + Γ model in RAxML. Hemimastigophora—including H. kukwesjijk and Spironema cf. multiciliatum from this study—are shown in red. Colours of other sequence names correspond to the same taxonomic groupings as in Fig. 3. The sequence of Spumella sp. strain BW2S, the prey for H. kukwesjijk, is included and marked with an asterisk. The numbers on branches show bootstrap percentages (1,000 replicates; values below 50% not shown). Branches in grey are half their original length. This tree was the reference phylogeny for pplacer analyses shown in Fig. 2. Scale bar denotes 0.1 expected substitutions per site.
Extended Data Fig. 4 Unrooted phylogeny of eukaryotes, 104 taxa dataset.
Phylogeny inferred from 351 genes, using maximum likelihood under the LG + C60 + F + Γ model. The numbers on branches show ultrafast bootstrap approximation percentages, with filled circles denoting 100% support. The Carpediemonas branch is shown reduced by 1/3 of the original length for display purposes. Scale bar denotes 0.1 expected substitutions per site.
Extended Data Fig. 5 Unrooted phylogeny using 58-nLB dataset.
Phylogeny inferred from 351 genes, using maximum likelihood under the LG + C60 + F + Γ model. The numbers on branches show PMSF bootstrap percentages (bootstrap support PMSF; 200 true bootstrap replicates), then ultrafast bootstrap approximation percentages (1,000 replicates). Filled circles denote 100% support with both methods. Scale bar denotes 0.1 expected substitutions per site.
Extended Data Fig. 6 Unrooted phylogeny using 58-nDP dataset.
Phylogeny inferred from 351 genes, using maximum likelihood under the LG + C60 + F + Γ model. The numbers on branches show PMSF bootstrap percentages (bootstrap support PMSF; 100 true bootstrap replicates), then ultrafast bootstrap approximation percentages (1,000 replicates). Filled circles denote 100% support with both methods. The branches leading to Bodo, Diplonema and Tetrahymena are shown reduced by 1/3. Scale bar denotes 0.1 expected substitutions per site.
Extended Data Fig. 7 Unrooted phylogeny using 61-SR4 dataset of 61 taxa.
Phylogeny inferred from 351 genes, with amino acids recoded as four states, using maximum likelihood under the GTR + R6 + F model. The numbers on branches show bootstrap percentages (500 true bootstrap replicates). Filled circles represent 100% support. The branches leading to Bodo, Diplonema and Tetrahymena are shown reduced by 1/3. Scale bar denotes 0.1 expected substitutions per site.
Extended Data Fig. 8 Summary of 61-SFSR analysis.
Chart follows the support for several important bipartitions with the sequential removal of the fastest-evolving sites from the 61-taxon, 351-gene dataset. The support values are ultra-fast bootstrap approximation percentages (1,000 replicates) inferred using maximum likelihood under the LG + C60 + F + Γ-derived PSMF model using a guide tree pruned of hemimastigotes (PMSF-nHEMI, see Methods); these values are not directly comparable to those from the other illustrated analyses.
Supplementary information
Supplementary Table 1
Full listing of environmental sequences attributable to Hemimastigophora, with habitat and location data.
Supplementary Table 2
Taxa used in phylogenomic analyses, organized by major group, with gene- and site-coverage statistics, and sources of data identified.
Supplementary Table 3
Genes of potential deep evolutionary significance in eukaryotes, searched for in the single-cell transcriptomes of Spironema and Hemimastix.
Rights and permissions
About this article
Cite this article
Lax, G., Eglit, Y., Eme, L. et al. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 564, 410–414 (2018). https://doi.org/10.1038/s41586-018-0708-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0708-8
Keywords
This article is cited by
-
Mitochondrial genomes revisited: why do different lineages retain different genes?
BMC Biology (2024)
-
The Pathology of the Brain Eating Amoeba Naegleria fowleri
Indian Journal of Microbiology (2024)
-
One high quality genome and two transcriptome datasets for new species of Mantamonas, a deep-branching eukaryote clade
Scientific Data (2023)
-
Combined nanometric and phylogenetic analysis of unique endocytic compartments in Giardia lamblia sheds light on the evolution of endocytosis in Metamonada
BMC Biology (2022)
-
Microbial predators form a new supergroup of eukaryotes
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.