Abstract
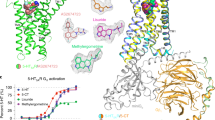
The 5-HT3A serotonin receptor1, a cationic pentameric ligand-gated ion channel (pLGIC), is the clinical target for management of nausea and vomiting associated with radiation and chemotherapies2. Upon binding, serotonin induces a global conformational change that encompasses the ligand-binding extracellular domain (ECD), the transmembrane domain (TMD) and the intracellular domain (ICD), the molecular details of which are unclear. Here we present two serotonin-bound structures of the full-length 5-HT3A receptor in distinct conformations at 3.32 Å and 3.89 Å resolution that reveal the mechanism underlying channel activation. In comparison to the apo 5-HT3A receptor, serotonin-bound states underwent a large twisting motion in the ECD and TMD, leading to the opening of a 165 Å permeation pathway. Notably, this motion results in the creation of lateral portals for ion permeation at the interface of the TMD and ICD. Combined with molecular dynamics simulations, these structures provide novel insights into conformational coupling across domains and functional modulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Coordinates of the 5-HT3A receptor structures have been deposited at the PDB under accession codes 6DG7 (state 1) and 6DG8 (state 2). The cryo-EM map has been deposited in the Electron Microscopy Data Bank under accession code EMD-7882 (state 1) and EMD-7883 (state 2). All relevant data are available from the authors.
References
Maricq, A. V., Peterson, A. S., Brake, A. J., Myers, R. M. & Julius, D. Primary structure and functional expression of the 5HT3 receptor, a serotonin-gated ion channel. Science 254, 432–437 (1991).
Machu, T. K. Therapeutics of 5-HT3 receptor antagonists: current uses and future directions. Pharmacol. Ther. 130, 338–347 (2011).
Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 346, 967–989 (2005).
Morales-Perez, C. L., Noviello, C. M. & Hibbs, R. E. X-ray structure of the human α4β2 nicotinic receptor. Nature 538, 411–415 (2016).
Du, J., Lu, W., Wu, S., Cheng, Y. & Gouaux, E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229 (2015).
Hassaine, G. et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281 (2014).
Miller, P. S. & Aricescu, A. R. Crystal structure of a human GABAA receptor. Nature 512, 270–275 (2014).
Connolly, C. N. Trafficking of 5-HT3 and GABAA receptors. Mol. Membr. Biol. 25, 293–301 (2008).
Peters, J. A. et al. The 5-hydroxytryptamine type 3 (5-HT3) receptor reveals a novel determinant of single-channel conductance. Biochem. Soc. Trans. 32, 547–552 (2004).
Baptista-Hon, D. T., Deeb, T. Z., Lambert, J. J., Peters, J. A. & Hales, T. G. The minimum M3–M4 loop length of neurotransmitter-activated pentameric receptors is critical for the structural integrity of cytoplasmic portals. J. Biol. Chem. 288, 21558–21568 (2013).
Basak, S. et al. Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat. Commun. 9, 514 (2018).
Marcus, Y. Ionic radii in aqueous solution. Chem. Rev. 88, 1475–1498 (1988).
Panicker, S., Cruz, H., Arrabit, C. & Slesinger, P. A. Evidence for a centrally located gate in the pore of a serotonin-gated ion channel. J. Neurosci. 22, 1629–1639 (2002).
Thompson, A. J. & Lummis, S. C. A single ring of charged amino acids at one end of the pore can control ion selectivity in the 5-HT3 receptor. Br. J. Pharmacol. 140, 359–365 (2003).
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001).
Kesters, D. et al. Structural basis of ligand recognition in 5-HT3 receptors. EMBO Rep. 14, 49–56 (2013).
Hansen, S. B. et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24, 3635–3646 (2005).
Beene, D. L. et al. Cation–π interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry 41, 10262–10269 (2002).
Yuan, S., Filipek, S. & Vogel, H. A gating mechanism of the serotonin 5-HT3 receptor. Structure 24, 816–825 (2016).
Sauguet, L. et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl Acad. Sci. USA 111, 966–971 (2014).
Miyazawa, A., Fujiyoshi, Y., Stowell, M. & Unwin, N. Nicotinic acetylcholine receptor at 4.6 Å resolution: transverse tunnels in the channel wall. J. Mol. Biol. 288, 765–786 (1999).
Kelley, S. P., Dunlop, J. I., Kirkness, E. F., Lambert, J. J. & Peters, J. A. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature 424, 321–324 (2003).
Hussy, N., Lukas, W. & Jones, K. A. Functional properties of a cloned 5-hydroxytryptamine ionotropic receptor subunit: comparison with native mouse receptors. J. Physiol. 481, 311–323 (1994).
McKinnon, N. K., Bali, M. & Akabas, M. H. Length and amino acid sequence of peptides substituted for the 5-HT3A receptor M3M4 loop may affect channel expression and desensitization. PLoS ONE 7, e35563 (2012).
Hales, T. G. et al. Common determinants of single channel conductance within the large cytoplasmic loop of 5-hydroxytryptamine type 3 and α4β2 nicotinic acetylcholine receptors. J. Biol. Chem. 281, 8062–8071 (2006).
Hu, X. Q., Sun, H., Peoples, R. W., Hong, R. & Zhang, L. An interaction involving an arginine residue in the cytoplasmic domain of the 5-HT3A receptor contributes to receptor desensitization mechanism. J. Biol. Chem. 281, 21781–21788 (2006).
Papke, D. & Grosman, C. The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J. Neurosci. 34, 7238–7252 (2014).
Yakel, J. L., Lagrutta, A., Adelman, J. P. & North, R. A. Single amino acid substitution affects desensitization of the 5-hydroxytryptamine type 3 receptor expressed in Xenopus oocytes. Proc. Natl Acad. Sci. USA 90, 5030–5033 (1993).
Revah, F. et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature 353, 846–849 (1991).
Basak, S., Schmandt, N., Gicheru, Y. & Chakrapani, S. Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel. eLife 6, e23886 (2017).
Panicker, S., Cruz, H., Arrabit, C., Suen, K. F. & Slesinger, P. A. Minimal structural rearrangement of the cytoplasmic pore during activation of the 5-HT3A receptor. J. Biol. Chem. 279, 28149–28158 (2004).
MacKenzie, D., Arendt, A., Hargrave, P., McDowell, J. H. & Molday, R. S. Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry 23, 6544–6549 (1984).
Stevens, R., Rusch, D., Solt, K., Raines, D. E. & Davies, P. A. Modulation of human 5-hydroxytryptamine type 3AB receptors by volatile anesthetics and n-alcohols. J. Pharmacol. Exp. Ther. 314, 338–345 (2005).
Thompson, A. J. & Lummis, S. C. A single channel mutation alters agonist efficacy at 5-HT3A and 5-HT3AB receptors. Br. J. Pharmacol. 170, 391–402 (2013).
Zhang, Z. & Chen, J. Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell 167, 1586–1597 (2016).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D 73, 496–502 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Collaborative Computational Project. N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).
Chen, V. B. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Chovancova, E. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput. Biol. 8, e1002708 (2012).
Stansfeld, P. J. & Sansom, M. S. Molecular simulation approaches to membrane proteins. Structure 19, 1562–1572 (2011).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Abraham, M. J., Murtola, T., Schulz, R., Pall, S. & Jeremy, C. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 14101 (2007).
Parrinello, M. R. A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Trick, J. L. et al. Functional annotation of ion channel structures by molecular simulation. Structure 24, 2207–2216 (2016).
Cordes, F. S., Bright, J. N. & Sansom, M. S. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323, 951–960 (2002).
Althoff, T., Hibbs, R. E., Banerjee, S. & Gouaux, E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014).
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011).
Acknowledgements
This research was supported in part by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research, and we thank them for the imaging time. We thank the Cleveland Center for Membrane and Structural Biology for the access to cryo-EM instrumentation, D. Major for assistance with hybridoma and cell culture at Department of Ophthalmology and Visual Sciences (NIH Core Grant P30EY11373), W. Boron for Xenopus oocytes and access to the oocyte rig, and G. Klesse and S. Tucker for the Channel Annotation Package methodology. This work was supported by an NIH grant (1R01GM108921), a cryo-EM supplement (3R01GM108921-03S1) to S.C. and an AHA postdoctoral Fellowship to S.B. (17POST33671152).
Reviewer information
Nature thanks S. M. Sine, A. I. Sobolevsky and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.B. and S.C. conceived the project and designed experimental procedures. S.B. carried out cryo-EM sample preparation, screening, data analysis and structure determination. Y.G. performed electrophysiological recordings. S.R. performed the molecular dynamics simulations under the supervision of M.S.P.S. S.C. supervised the execution of the experiments, data analysis and interpretation. S.B., Y.G. and S.C. drafted the manuscript. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Data processing workflow.
a, A representative micrograph of 5-HT3A receptor incubated with 100 μM serotonin in vitreous ice (top). Selected 2D classes showing various orientations (bottom). b, A schematic of the steps followed in data processing leading to 3.32 Å and 3.89 Å reconstructions of state 1 and state 2, respectively. Each subunit is shown in a different colour for clarity. Classes within the boxes represent two distinct conformations. Based on the number of particles for each state, it appears that state 1 is more populated than state 2 under our current conditions.
Extended Data Fig. 2 Estimation of resolution and validation of the models.
a, FSC curves before (red) and after (blue) the application of soft mask in RELION for state 1 (left) and state 2 (right). The dashed line represents FSC of 0.143. b, For cross validation, FSC curves of the refined model versus summed map (full dataset, blue), refined model versus half map 1 (used during refinement, cyan), and refined model versus half map 2 (not used during refinement, purple) were calculated for state 1 (left) and state 2 (right). c, Local resolution of state 1 (left) and state 2 (right) reconstructions were estimated using the ResMap program40.
Extended Data Fig. 3 Map correlation of state 1 and state 2.
Various regions of the model (shown as a cartoon) and corresponding density map (mesh) around the residues are shown to validate the final model. Residues are depicted as sticks. The depicted regions in state 1 and the corresponding contour levels: Cys loop (7.0σ), loop C (8.0σ), loop F (8.0σ), M2 (6.0σ), M2–M3 linker (7.0σ), M4 (6.5σ), MX helix (7.5σ) and MA helix (7.5σ). The depicted regions in state 2 and the corresponding contour levels: Cys loop (8.5σ), loop C (8.0σ), loop F (8.0σ), M2 (6.0σ), M2–M3 linker (7.0σ), M4 (7.0σ), MX helix (7.0σ) and MA helix (6.0σ).
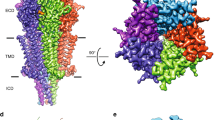
Extended Data Fig. 4 Serotonin-induced conformational changes in the ECD and TMD.
a, A global alignment of the apo structure with state 1 (left) and state 2 (right). The top panel shows the ECD and the bottom panel shows the TMD, both viewed from the extracellular end. The arrows indicate the direction of rotation with respect to the apo state. State 1 and state 2 structures superimpose with the apo ECD with a root mean square deviation (r.m.s.d.) of 1.16 for state 1 and 1.41 for state 2 (residues 8–220). State 1, and particularly state 2, diverge markedly in the TMD and ICD (r.m.s.d. of 1.1 for state 1 and 4.24 for state 2 for residues 221–462). b, A side-view of the ECDs upon aligning state 1 and state 2 to the apo state. c, A top view of the TMDs when aligned with respect to the ECD. The arrows show relative displacements in different regions of state 1 and state 2 with respect to the apo structure.
Extended Data Fig. 5 Inter-subunit interaction at the ECD–TMD–ICD interface.
a, Inter-subunit interactions at the ECD–TMD interface in the apo state, state 1 and state 2. b, Inter-subunit interactions at the TMD–ICD interface in the three states. The potential interactions were predicted as polar contacts in PyMOL. Interacting residues are shown as sticks. Residue labels are colour-coded based on their location. The apo state has the largest buried surface area (31,610 Å2) which progressively decreases in state 1 (30,960 Å2) and then state 2 (25,340 Å2).
Extended Data Fig. 6 The intracellular domain of state 2.
a, A detailed view of the ICD with key residues shown in stick representation. Only two adjacent subunits are shown for clarity. The solvent-accessible vestibule in the ICD calculated using Caver3.047 with a minimum cavity radius of 2.8 Å is shown as dark-cyan spheres. The positively charged residues lining the portal are shown as blue sticks. The negatively charged residues in the vicinity are shown in red-brown. Residues that form the hydrophobic patch at the N-terminal end of the MA helix are shown as green sticks. Residues His309 (post-M3 loop) and Glu250 (M2) are in a potential interaction and are shown in magenta. b, A zoomed view of the ICD to highlight the break in MA–M4 helices (highlighted in magenta) at Gly430. Glycine-mediated transmembrane-helix distortion at the i−3 position is well-studied58, and Gly430 may have a dynamic role at the hinge point between MA and M4 helices. A similar bend in the MA–M4 helix was previously observed in the Torpedo marmorata nAChR structure even in the absence of glycine at the equivalent position3.
Extended Data Fig. 7 Molecular dynamics simulations of state 1.
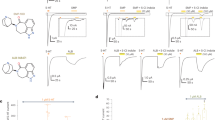
a, Trajectories of water and Na+ ion coordinates within 5 Å of the channel axis inside the pore over 100 ns with a 0.2-V transmembrane potential difference, with the cytoplasmic side having a positive potential. Stretches of white regions indicate areas devoid of water or ions. b, Time-averaged radii along the central pore axis of the state 1 structure during two 10-ns fractions of the simulation (within the boxed region of a) (top). The orange arrows indicate positions of Leu260. The dashed line indicates the approximate radius of a hydrated Na+ ion. Free-energy profile of a water molecule along the central pore axis during the 10-ns window (bottom). The barrier at the Leu260 position disappears in the 45–55-ns time frame. c, Snapshots of pore conformation around Leu260 (shown in stick representation) during the corresponding time window. The widening of the pore radii and the disappearance of the barrier for water permeation is associated with the rotameric reorientation of the Leu260 side chain (indicated by the arrow). d, An overlay of the pore radii from the two time windows. Changes at position Leu260 are marked by the arrow. e, Snapshot of pore conformation around Leu260 as a Na+ ion (purple) is passing through, with two Leu260 side chains rotated away (indicated by *). Three independent simulations were run.
Extended Data Fig. 8 Snapshots of the state 1 pore conformation from the molecular dynamics simulation.
a, Side-chain orientations of Leu260 and Glu250 at different points during the simulation (indicated in Extended Data Fig. 7). b, The corresponding pore radius profiles. The positions of Glu250 and Leu260 are highlighted.
Extended Data Fig. 9 Comparison of pLGIC pore profiles.
Pore profiles calculated using the HOLE program46 for the M2 region of nAChR (PDB ID: 5KXI)4, GABAA receptor β3 homopentamer (PDB ID: 4COF)7, glutamate-gated chloride channel (GluCl) (apo structure, PDB ID: 4TNV59; ivermectin-bound, PDB ID: 3RHW60), glycine receptor (GlyR) (strychnine-bound, PDB ID: 3JAD; glycine-bound, PDB ID: 3JAE; glycine- and ivermectin-bound, PDB ID: 3JAF)5 and 5-HT3A receptor (apo structure, PDB ID: 6BE111; state 1, PDB ID: 6DG7; state 2, PDB ID: 6DG8). Only two M2 helices are shown as ribbon, for clarity. Pore-facing residues are shown as stick representation. Green and magenta spheres define radii of 1.8–3.3 Å and >3.3 Å, respectively.
Supplementary information
Video 1: Ion permeation in State 2
Trajectory of a single hydrated Na+ ion (purple) entering from the ECD and exiting through the ICD portals (-200 mV). The video corresponds to 35 ns of a 100 ns simulation. Periodic boundaries of the simulation cell are indicated by the solid frame. The Na+ ion and surrounding water molecules (blue) within 4 Å of it are shown. Other ions and water molecules are omitted for clarity. Leu260 is coloured red. Ions passage through the lateral gates were observed in multiple simulations (100 ns replicates at 0.7 M NaCl, 200 mV). In all 23 events where we see a full (i.e. bulk to bulk) conductance event, the ion passes through the lateral gates.
Rights and permissions
About this article
Cite this article
Basak, S., Gicheru, Y., Rao, S. et al. Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274 (2018). https://doi.org/10.1038/s41586-018-0660-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0660-7
Keywords
This article is cited by
-
Structural basis for partial agonism in 5-HT3A receptors
Nature Structural & Molecular Biology (2024)
-
Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment
Nature Communications (2024)
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
Structural determinants for activity of the antidepressant vortioxetine at human and rodent 5-HT3 receptors
Nature Structural & Molecular Biology (2024)
-
Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.