Abstract
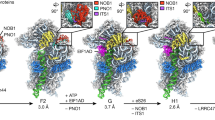
The formation of eukaryotic ribosomal subunits extends from the nucleolus to the cytoplasm and entails hundreds of assembly factors. Despite differences in the pathways of ribosome formation, high-resolution structural information has been available only from fungi. Here we present cryo-electron microscopy structures of late-stage human 40S assembly intermediates, representing one state reconstituted in vitro and five native states that range from nuclear to late cytoplasmic. The earliest particles reveal the position of the biogenesis factor RRP12 and distinct immature rRNA conformations that accompany the formation of the 40S subunit head. Molecular models of the late-acting assembly factors TSR1, RIOK1, RIOK2, ENP1, LTV1, PNO1 and NOB1 provide mechanistic details that underlie their contribution to a sequential 40S subunit assembly. The NOB1 architecture displays an inactive nuclease conformation that requires rearrangement of the PNO1-bound 3′ rRNA, thereby coordinating the final rRNA folding steps with site 3 cleavage.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Woolford, J. L. Jr & Baserga, S. J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Zemp, I. & Kutay, U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581, 2783–2793 (2007).
Tschochner, H. & Hurt, E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13, 255–263 (2003).
Phipps, K. R., Charette, J. & Baserga, S. J. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA 2, 1–21 (2011).
Rouquette, J., Choesmel, V. & Gleizes, P. E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 24, 2862–2872 (2005).
Wyler, E. et al. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA 17, 189–200 (2011).
Widmann, B. et al. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 23, 22–35 (2012).
Zorbas, C. et al. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 26, 2080–2095 (2015).
Heuer, A. et al. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. eLife 6, e30189 (2017).
Scaiola, A. et al. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 37, e98499 (2018).
Henras, A. K., Plisson-Chastang, C., O’Donohue, M. F., Chakraborty, A. & Gleizes, P. E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242 (2015).
Tafforeau, L. et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol. Cell 51, 539–551 (2013).
Badertscher, L. et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Reports 13, 2879–2891 (2015).
Larburu, N. et al. Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res. 44, 8465–8478 (2016).
Oeffinger, M., Dlakic, M. & Tollervey, D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18, 196–209 (2004).
McCaughan, U. M. et al. Pre-40S ribosome biogenesis factor Tsr1 is an inactive structural mimic of translational GTPases. Nat. Commun. 7, 11789 (2016).
.Sun, Q. et al. Molecular architecture of the 90S small subunit pre-ribosome. eLife 6, (2017).
Strunk, B. S. et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333, 1449–1453 (2011).
Ferreira-Cerca, S., Kiburu, I., Thomson, E., LaRonde, N. & Hurt, E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 42, 8635–8647 (2014).
Woolls, H. A., Lamanna, A. C. & Karbstein, K. Roles of Dim2 in ribosome assembly. J. Biol. Chem. 286, 2578–2586 (2011).
Turowski, T. W. et al. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 42, 12189–12199 (2014).
Strunk, B. S., Novak, M. N., Young, C. L. & Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150, 111–121 (2012).
Glatter, T., Wepf, A., Aebersold, R. & Gstaiger, M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
The authors thank S. Rieder, H. Sieber and A. Gilmozzi for technical assistance, T. Fröhlich for mass-spectrometry analysis and E. Hurt, T. Becker and L. Kater for discussions and critical comments on the manuscript. This research was supported by grants from the Deutsche Forschungsgemeinschaft (SFB646, GRK1721 and FOR1805 to R.B.) and by an European Research Council (ERC) Advanced Grant (CRYOTRANSLATION) to R.B. M.A. is supported by a DFG fellowship through the Graduate School of Quantitative Biosciences Munich (QBM).
Author information
Authors and Affiliations
Contributions
M.A., J.C., O.B. and R.B. designed the study. M.A. generated stable cell lines and purified native complexes. J.C. cloned and purified biogenesis factors and conducted the reconstitution. M.A., J.C. and O.B. prepared the cryo-EM samples and O.B. collected cryo-EM data. M.A. and J.C. processed the cryo-EM data for their respective samples and J.C. built the molecular models with the help of M.A. M.A., J.C. and R.B. analysed the structures, interpreted results and all authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
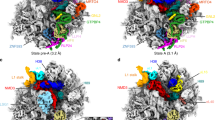
Extended Data Fig. 1 Sample preparation and cryo-EM analysis of the pre-40S ribosome.
a, Coomassie stained SDS–PAGE analysis of samples used in this work with bands labelled as identified by mass spectrometry. RBFs that we do not observe in our structures are marked with an asterisk. 60S bands are shown for comparison (see Supplementary Fig. 1 for source data). b, Representative micrographs from three datasets, low-pass filtered at 15 Å. c, Representative 2D classes of three datasets showing various orientations of pre-40S particles. d, Cryo-EM data processing scheme for all datasets with final volumes highlighted in red. Classes that could not be further refined or sorted are labelled flexible or noisy. For PNO1 N-TAP, two datasets were collected and individually processed. Particles of respective states were combined and further analysed (see Methods). Complex purification was done three times independently with the same results. Cryo-EM data collection and analysis was done twice for native complexes with similar results. Mass spectrometry analysis, pre-40S reconstitution and data collection for datasets 3 and 4 were done once. DS, dataset.
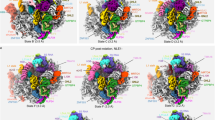
Extended Data Fig. 2 Local resolution, refinement and model statistics.
a, Local resolution distribution as estimated by Relion ranging from approximately 3 Å in well-resolved areas to 12 Å in more flexible parts. Colouring according to scale bars. b, Fourier shell correlation (FSC) plot with average resolutions according to the ‘gold standard’ (FSC = 0.143) stated in the legend. c, FSC plot of the state C model against cryo-EM maps as calculated by REFMAC5 (see Supplementary Data 1 for source data). d, Structural comparison between state C and a pre-40S particle from yeast (PDB code 6EML). Assembly factors coloured as in Fig. 1a. e, Cartoon representations of models of RBFs (top) are shown together with their respective density (mid). Volumes are coloured according to local resolution, which ranges from 3 Å in more rigid areas to 9 Å in flexible parts. RRP12, BUD23 and TRMT112 are less resolved, which only permitted placing of dummy helices and rigid body fitting of homology models, respectively. Examples of well resolved areas are depicted below.
Extended Data Fig. 3 Structural details of assembly factors and beak formation.
a, Positioning of RRP12 in state B shows interaction with eS17 and uS9, as well as rRNA h34, h39, h40 and h41. Two additional helices of unknown identity bridge RRP12 and the head. Poly-alanine helices are placed in respective densities. b, Recruitment of NOB1 and the uS2–uS5–eS21 cluster after h35–h37 flipping; eS17 is omitted from this view. c, Close-up view of the beak region before (left) and after (right) replacement of ENP1-LTV1 with the uS3–eS10–uS20 cluster. c, Movement of h34 during maturation.
Extended Data Fig.4 The maturation of the decoding centre.
a, Close-up views of the decoding centre in density (top) and model (bottom) representation showing the position of the biogenesis factors close to h44. b, Domain arrangement of TSR1. Insertions and extensions of TSR1 are labelled. c, Conformational change of the N terminus of eS31 during maturation. The alignment was based on the C-terminal zinc-finger domain. d, Overall structure of the reconstituted particle low-pass filtered at 12 Å, showing the approximate positioning of RIOK1 (blue).
Extended Data Fig. 5 PNO1, NOB1 and coordination of the 3′ rRNA end.
a, The 3′ end region (box) throughout the maturation process. PNO1 and NOB1 bind similarly in state B–E and R. b, Cartoon representation of eS26 and PNO1 with parts of the pre-18S rRNA at different stages in maturation. h44 is shifted in state C. Displacement of eS26 by PNO1 in state R leads to a partially shifted h44. c, Overall structure of PNO1 with its two KH domains. d, e, Detailed view on the effects of PNO1 binding. Clashing of PNO1 with h28 and the linker region between h44 and h45 leads to a disruption of the base stacking between G1207 and G1837 (d) and A1835 and A1863 (e). f, Residues involved in novel base pairing with their respective electron density in state C. g, Comparison of the PIN domain of human (hs) NOB1 in state C and the NMR structure of Nob1 from Pyrococcus horikoshii (ph; PDB code 2LCQ), with their conserved active site residues highlighted. h, Summary of residues involved in binding of the rRNA 3′ end (asterisk indicates stacking). Electron density of state C surrounding the 3′ end is shown.
Extended Data Fig. 6 2D diagram of the pre-18S rRNA head region of state C.
Extended secondary structure diagram of the pre-18S rRNA head region of state C. Residues mentioned throughout the text are highlighted. Related to Fig. 4b.
Extended Data Fig. 7 60S subunits components clash with immature h44.
Overview over clash sites of large subunit components with pre-40S particles. Left, initiation factor eIF5B, which is potentially involved in formation of the 80S-like ribosome complex, occupies a similar binding site as TSR1. Middle, ribosomal protein uL23 and helices H69 and H71 of 28S rRNA clash with h44 in its immature position. Right, finally helix H66 of 28S rRNA clashes with a RIOK1 helix. A model of an 80S ribosome with eIF5B (PDB code 4UJD) was aligned to pre-40S states, and the factors are shown as overlays together with one of the 40S precursors.
Supplementary information
Supplementary Information
This figure shows the uncropped SDS-PAGE images in Extended Data Figure 1a. The black rectangles represent the sections shown.
Source Data
Rights and permissions
About this article
Cite this article
Ameismeier, M., Cheng, J., Berninghausen, O. et al. Visualizing late states of human 40S ribosomal subunit maturation. Nature 558, 249–253 (2018). https://doi.org/10.1038/s41586-018-0193-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0193-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.