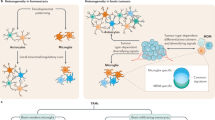
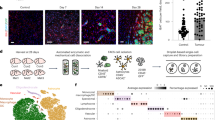
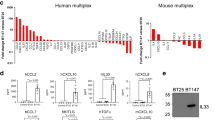
Abstract
Microglia are the resident innate immune cells of the immune-privileged CNS and, as such, represent the first line of defence against tissue injury and infection. Given their location, microglia are undoubtedly the first immune cells to encounter a developing primary brain tumour. Our knowledge of these cells is therefore important to consider in the context of such neoplasms. As the heterogeneous nature of the most aggressive primary brain tumours is thought to underlie their poor prognosis, this Review places a special emphasis on the heterogeneity of the tumour-associated microglia and macrophage populations present in primary brain tumours. Where available, specific information on microglial heterogeneity in various types and subtypes of brain tumour is included. Emerging evidence that highlights the importance of considering the heterogeneity of both the tumour and of microglial populations in providing improved treatment outcomes for patients is also discussed.
Key points
-
Brain tumours are heterogeneous and can now be subdivided into molecular subtypes based on genetic alterations.
-
These tumour subtypes contain tumour-associated microglia and/or macrophages that can vary in number and phenotype.
-
Microglia display acquired as well as intrinsic spatiotemporal heterogeneity (that is, reaction states and subtypes) that should be considered in the context of the tumour microenvironment.
-
Understanding the complexity of tumour-associated microglia and macrophage populations could have diagnostic value and generate novel avenues for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Spittau, B. Aging microglia-phenotypes, functions and implications for age-related neurodegenerative diseases. Front. Aging Neurosci. 9, 194 (2017).
Wolf, S. A., Boddeke, H. W. & Kettenmann, H. Microglia in physiology and disease. Annu. Rev. Physiol. 79, 619–643 (2017).
Li, Q. & Barres, B. A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18, 225–242 (2018).
Ostrom, Q. T. et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 16, 896–913 (2014).
Bondy, M. L. et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer 113, 1953–1968 (2008).
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Preusser, M., Brastianos, P. K. & Mawrin, C. Advances in meningioma genetics: novel therapeutic opportunities. Nat. Rev. Neurol. 14, 106–115 (2018).
Villa, A. et al. Sex-specific features of microglia from adult mice. Cell Rep. 23, 3501–3511 (2018).
Guneykaya, D. et al. Transcriptional and translational differences of microglia from male and female brains. Cell Rep. 24, 2773–2783 (2018).
Kodama, L. et al. Microglial microRNAs mediate sex-specific responses to tau pathology. Nat. Neurosci. 23, 167–171 (2020).
Ochocka, N. et al. Single-cell RNA sequencing reveals functional heterogeneity and sex differences of glioma-associated brain macrophages. Preprint at bioRxiv https://doi.org/10.1101/752949 (2020).
McKinney, P. A. Brain tumours: incidence, survival, and aetiology. J. Neurol. Neurosurg. Psychiatry 75 (Suppl. 2), ii12–ii17 (2004).
Jessa, S. et al. Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 51, 1702–1713 (2019).
Masuda, T., Sankowski, R., Staszewski, O. & Prinz, M. Microglia heterogeneity in the single-cell era. Cell Rep. 30, 1271–1281 (2020).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019).
Bisht, K. et al. Dark microglia: a new phenotype predominantly associated with pathological states. Glia 64, 826–839 (2016).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019).
Lenz, K. M. & Nelson, L. H. Microglia and beyond: innate immune cells as regulators of brain development and behavioral function. Front. Immunol. 9, 698 (2018).
Grabert, K. et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19, 504–516 (2016).
Stratoulias, V., Venero, J. L., Tremblay, M. E. & Joseph, B. Microglial subtypes: diversity within the microglial community. EMBO J. 38, e101997 (2019).
Wlodarczyk, A. et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308 (2017).
Ueno, M. et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 16, 543–551 (2013).
Benmamar-Badel, A., Owens, T. & Wlodarczyk, A. Protective microglial subset in development, aging, and disease: lessons from transcriptomic studies. Front. Immunol. 11, 430 (2020).
Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019).
Yao, M. et al. Astrocytic trans-differentiation completes a multicellular paracrine feedback loop required for medulloblastoma tumor growth. Cell 180, 502–520 (2020).
Gibson, P. et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099 (2010).
de Pablo, F. & de la Rosa, E. J. The developing CNS: a scenario for the action of proinsulin, insulin and insulin-like growth factors. Trends Neurosci. 18, 143–150 (1995).
Svalina, M. N. et al. IGF1R as a key target in high risk, metastatic medulloblastoma. Sci. Rep. 6, 27012 (2016).
Sato-Hashimoto, M. et al. Microglial SIRPα regulates the emergence of CD11c+ microglia and demyelination damage in white matter. eLife 8, e42025 (2019).
Van Vuurden, D. G. H. E. et al. SIRPα is transcriptionally downregulated by epigenetic silencing in medulloblastoma. J. Mol. Clin. Med. 1, 157–168 (2018).
Gholamin, S. et al. Disrupting the CD47–SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl Med. 9, eaaf2968 (2017).
Szulzewsky, F. et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS ONE 10, e0116644 (2015).
Domingues, P. H. et al. Association between inflammatory infiltrates and isolated monosomy 22/del(22q) in meningiomas. PLoS ONE 8, e74798 (2013).
Bertolotto, A., Caterson, B., Canavese, G., Migheli, A. & Schiffer, D. Monoclonal antibodies to keratan sulfate immunolocalize ramified microglia in paraffin and cryostat sections of rat brain. J. Histochem. Cytochem. 41, 481–487 (1993).
Bertolotto, A., Agresti, C., Castello, A., Manzardo, E. & Riccio, A. 5D4 keratan sulfate epitope identifies a subset of ramified microglia in normal central nervous system parenchyma. J. Neuroimmunol. 85, 69–77 (1998).
Wilms, H., Wollmer, M. A. & Sievers, J. In vitro-staining specificity of the antibody 5-D-4 for microglia but not for monocytes and macrophages indicates that microglia are a unique subgroup of the myelomonocytic lineage. J. Neuroimmunol. 98, 89–95 (1999).
Jones, L. L. & Tuszynski, M. H. Spinal cord injury elicits expression of keratan sulfate proteoglycans by macrophages, reactive microglia, and oligodendrocyte progenitors. J. Neurosci. 22, 4611–4624 (2002).
Zhang, Z. et al. Deficiency of a sulfotransferase for sialic acid-modified glycans mitigates Alzheimer’s pathology. Proc. Natl Acad. Sci. USA 114, E2947–E2954 (2017).
Hirano, K. et al. Ablation of keratan sulfate accelerates early phase pathogenesis of ALS. PLoS ONE 8, e66969 (2013).
Mughal, A. A. et al. Patterns of invasive growth in malignant gliomas-the hippocampus emerges as an invasion-spared brain region. Neoplasia 20, 643–656 (2018).
Lapin, D. H., Tsoli, M. & Ziegler, D. S. Genomic insights into diffuse intrinsic pontine glioma. Front. Oncol. 7, 57 (2017).
Vitanza, N. A. & Monje, M. Diffuse intrinsic pontine glioma: from diagnosis to next-generation clinical trials. Curr. Treat. Options Neurol. 21, 37 (2019).
Nioka, H., Matsumura, K., Nakasu, S. & Handa, J. Immunohistochemical localization of glycosaminoglycans in experimental rat glioma models. J. Neurooncol. 21, 233–242 (1994).
Kato, Y. et al. Increased expression of highly sulfated keratan sulfate synthesized in malignant astrocytic tumors. Biochem. Biophys. Res. Commun. 369, 1041–1046 (2008).
Leiphrakpam, P. D. et al. Role of keratan sulfate expression in human pancreatic cancer malignancy. Sci. Rep. 9, 9665 (2019).
Yin, J. et al. Transforming growth factor-β1 upregulates keratan sulfate and chondroitin sulfate biosynthesis in microglias after brain injury. Brain Res. 1263, 10–22 (2009).
Lin, G. L. et al. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol. Commun. 6, 51 (2018).
Jander, S., Schroeter, M., Fischer, J. & Stoll, G. Differential regulation of microglial keratan sulfate immunoreactivity by proinflammatory cytokines and colony-stimulating factors. Glia 30, 401–410 (2000).
Lun, M. P. et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci. 35, 4903–4916 (2015).
Wolff, J. E., Sajedi, M., Brant, R., Coppes, M. J. & Egeler, R. M. Choroid plexus tumours. Br. J. Cancer 87, 1086–1091 (2002).
Chen, S. K. et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785 (2010).
De, S. et al. Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 145, dev152306 (2018).
Tremblay, M. E., Lowery, R. L. & Majewska, A. K. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 8, e1000527 (2010).
Wake, H., Moorhouse, A. J., Jinno, S., Kohsaka, S. & Nabekura, J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 29, 3974–3980 (2009).
Cunningham, C. L., Martinez-Cerdeno, V. & Noctor, S. C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33, 4216–4233 (2013).
Burns, J. C. et al. Differential accumulation of storage bodies with aging defines discrete subsets of microglia in the healthy brain. eLife 9, e57495 (2020).
Sedgwick, J. D. et al. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc. Natl Acad. Sci. USA 88, 7438–7442 (1991).
Muller, A., Brandenburg, S., Turkowski, K., Muller, S. & Vajkoczy, P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int. J. Cancer 137, 278–288 (2015).
Bowman, R. L. et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 17, 2445–2459 (2016).
Yu, K. et al. Surveying brain tumor heterogeneity by single-cell RNA-sequencing of multi-sector biopsies. Natl Sci. Rev. 7, 1306–1318 (2020).
Muller, S. et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18, 234 (2017).
Hambardzumyan, D., Gutmann, D. H. & Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27 (2016).
Charles, N. A., Holland, E. C., Gilbertson, R., Glass, R. & Kettenmann, H. The brain tumor microenvironment. Glia 60, 502–514 (2012).
Wei, J., Gabrusiewicz, K. & Heimberger, A. The controversial role of microglia in malignant gliomas. Clin. Dev. Immunol. 2013, 285246 (2013).
Gutmann, D. H. & Kettenmann, H. Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron 104, 442–449 (2019).
Haage, V. et al. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol. Commun. 7, 20 (2019).
Louis, D. N. et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109 (2007).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Verhaak, R. G. W. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Houillier, C. et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75, 1560–1566 (2010).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Wang, Q. et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56 (2017).
Behnan, J., Finocchiaro, G. & Hanna, G. The landscape of the mesenchymal signature in brain tumours. Brain 142, 847–866 (2019).
Engler, J. R. et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS ONE 7, e43339 (2012).
Shan, X. et al. Prognostic value of a nine-gene signature in glioma patients based on tumor-associated macrophages expression profiling. Clin. Immunol. 216, 108430 (2020).
Kaffes, I. et al. Human mesenchymal glioblastomas are characterized by an increased immune cell presence compared to proneural and classical tumors. Oncoimmunology 8, e1655360 (2019).
Darmanis, S. et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017).
Nduom, E. K., Weller, M. & Heimberger, A. B. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol. 17 (Suppl. 7), vii9–vii14 (2015).
Watters, J. J., Schartner, J. M. & Badie, B. Microglia function in brain tumors. J. Neurosci. Res. 81, 447–455 (2005).
Gieryng, A., Pszczolkowska, D., Walentynowicz, K. A., Rajan, W. D. & Kaminska, B. Immune microenvironment of gliomas. Lab. Invest. 97, 498–518 (2017).
Walentynowicz, K. A. et al. In search of reliable markers for glioma-induced polarization of microglia. Front. Immunol. 9, 1329 (2018).
Chen, Z. et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 77, 2266–2278 (2017).
Klemm, F. et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660 (2020).
Friebel, E. et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642 (2020).
Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006).
Northcott, P. A. et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011).
Thompson, M. C. et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 24, 1924–1931 (2006).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754.e6 (2017).
Griesinger, A. M. et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J. Immunol. 191, 4880–4888 (2013).
Pham, C. D. et al. Differential immune microenvironments and response to immune checkpoint blockade among molecular subtypes of murine medulloblastoma. Clin. Cancer Res. 22, 582–595 (2016).
Bockmayr, M. et al. Subgroup-specific immune and stromal microenvironment in medulloblastoma. Oncoimmunology 7, e1462430 (2018).
Maximov, V. et al. Tumour-associated macrophages exhibit anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat. Commun. 10, 2410 (2019).
Lee, C. et al. M1 macrophage recruitment correlates with worse outcome in SHH medulloblastomas. BMC Cancer 18, 535 (2018).
Seizinger, B. R., de la Monte, S., Atkins, L., Gusella, J. F. & Martuza, R. L. Molecular genetic approach to human meningioma: loss of genes on chromosome 22. Proc. Natl Acad. Sci. USA 84, 5419–5423 (1987).
Sanson, M. et al. Germline deletion in a neurofibromatosis type 2 kindred inactivates the NF2 gene and a candidate meningioma locus. Hum. Mol. Genet. 2, 1215–1220 (1993).
Brastianos, P. K. et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 45, 285–289 (2013).
Sahm, F. et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 18, 682–694 (2017).
Wood, G. W. & Morantz, R. A. Immunohistologic evaluation of the lymphoreticular infiltrate of human central nervous system tumors. J. Natl Cancer Inst. 62, 485–491 (1979).
Rossi, M. L., Cruz Sanchez, F., Hughes, J. T., Esiri, M. M. & Coakham, H. B. Immunocytochemical study of the cellular immune response in meningiomas. J. Clin. Pathol. 41, 314–319 (1988).
Bo, L., Mork, S. J. & Nyland, H. An immunohistochemical study of mononuclear cells in meningiomas. Neuropathol. Appl. Neurobiol. 18, 548–558 (1992).
Asai, J. et al. Fluorescence automatic cell sorter and immunohistochemical investigation of CD68-positive cells in meningioma. Clin. Neurol. Neurosurg. 101, 229–234 (1999).
Strik, H. M., Stoll, M. & Meyermann, R. Immune cell infiltration of intrinsic and metastatic intracranial tumours. Anticancer Res. 24, 37–42 (2004).
Grund, S. et al. The microglial/macrophagic response at the tumour-brain border of invasive meningiomas. Neuropathol. Appl. Neurobiol. 35, 82–88 (2009).
Proctor, D. T. et al. Tumor-associated macrophage infiltration in meningioma. Neurooncol. Adv. 1, vdz018 (2019).
Adams, C. L. et al. A rapid robust method for subgrouping non-NF2 meningiomas according to genotype and detection of lower levels of M2 macrophages in AKT1 E17K mutated tumours. Int. J. Mol. Sci. 21, 1273 (2020).
Lauber, C., Klink, B. & Seifert, M. Comparative analysis of histologically classified oligodendrogliomas reveals characteristic molecular differences between subgroups. BMC Cancer 18, 399 (2018).
Kamoun, A. et al. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat. Commun. 7, 11263 (2016).
Deininger, M. H. et al. Cyclooxygenase (COX)-1 expressing macrophages/microglial cells and COX-2 expressing astrocytes accumulate during oligodendroglioma progression. Brain Res. 885, 111–116 (2000).
Deininger, M. H. et al. Heme oxygenase (HO)-1 expressing macrophages/microglial cells accumulate during oligodendroglioma progression. Brain Res. 882, 1–8 (2000).
Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017).
Nam, S. J. et al. Tumor-infiltrating immune cell subpopulations and programmed death ligand 1 (PD-L1) expression associated with clinicopathological and prognostic parameters in ependymoma. Cancer Immunol. Immunother. 68, 305–318 (2019).
Pajtler, K. W. et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27, 728–743 (2015).
Lester, A. & McDonald, K. L. Intracranial ependymomas: molecular insights and translation to treatment. Brain Pathol. 30, 3–12 (2020).
Eder, N. et al. YAP1/TAZ drives ependymoma-like tumour formation in mice. Nat. Commun. 11, 2380 (2020).
Liu, S. J. et al. Multiplatform molecular profiling reveals epigenomic intratumor heterogeneity in ependymoma. Cell Rep. 30, 1300–1309.e5 (2020).
Elsarrag, M., Patel, P. D., Chatrath, A., Taylor, D. & Jane, J. A. Genomic and molecular characterization of pituitary adenoma pathogenesis: review and translational opportunities. Neurosurg. Focus. 48, E11 (2020).
Lu, J. Q. et al. Immune cell infiltrates in pituitary adenomas: more macrophages in larger adenomas and more T cells in growth hormone adenomas. Endocr. Pathol. 26, 263–272 (2015).
Yagnik, G., Rutowski, M. J., Shah, S. S. & Aghi, M. K. Stratifying nonfunctional pituitary adenomas into two groups distinguished by macrophage subtypes. Oncotarget 10, 2212–2223 (2019).
Sato, M. et al. Analysis of tumor angiogenesis and immune microenvironment in non-functional pituitary endocrine tumors. J. Clin. Med. 8, 695 (2019).
Desbaillets, I. et al. Human astrocytomas and glioblastomas express monocyte chemoattractant protein-1 (MCP-1) in vivo and in vitro. Int. J. Cancer 58, 240–247 (1994).
Chang, A. L. et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 76, 5671–5682 (2016).
Lindemann, C., Marschall, V., Weigert, A., Klingebiel, T. & Fulda, S. SMAC mimetic-induced upregulation of CCL2/MCP-1 triggers migration and invasion of glioblastoma cells and influences the tumor microenvironment in a paracrine manner. Neoplasia 17, 481–489 (2015).
Platten, M. et al. Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann. Neurol. 54, 388–392 (2003).
Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018).
Brana, I. et al. Carlumab, an anti-C-C chemokine ligand 2 monoclonal antibody, in combination with four chemotherapy regimens for the treatment of patients with solid tumors: an open-label, multicenter phase 1b study. Target. Oncol. 10, 111–123 (2015).
Elmore, M. R. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Easley-Neal, C., Foreman, O., Sharma, N., Zarrin, A. A. & Weimer, R. M. CSF1R ligands IL-34 and CSF1 are differentially required for microglia development and maintenance in white and gray matter brain regions. Front. Immunol. 10, 2199 (2019).
Pyonteck, S. M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013).
Quail, D. F. et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 352, aad3018 (2016).
Butowski, N. et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol. 18, 557–564 (2016).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02526017 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04257617 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02216409 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03512340 (2020).
Kosaka, A., Ohkuri, T. & Okada, H. Combination of an agonistic anti-CD40 monoclonal antibody and the COX-2 inhibitor celecoxib induces anti-glioma effects by promotion of type-1 immunity in myeloid cells and T-cells. Cancer Immunol. Immunother. 63, 847–857 (2014).
Shoji, T. et al. Local convection-enhanced delivery of an anti-CD40 agonistic monoclonal antibody induces antitumor effects in mouse glioma models. Neuro Oncol. 18, 1120–1128 (2016).
Vonderheide, R. H. et al. Phase I study of the CD40 agonist antibody CP-870,893 combined with carboplatin and paclitaxel in patients with advanced solid tumors. Oncoimmunology 2, e23033 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03389802 (2020).
Stathopoulos, A. et al. Development of immune memory to glial brain tumors after tumor regression induced by immunotherapeutic Toll-like receptor 7/8 activation. Oncoimmunology 1, 298–305 (2012).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01204684. (2020).
Gupta, K. & Burns, T. C. Radiation-induced alterations in the recurrent glioblastoma microenvironment: therapeutic implications. Front. Oncol. 8, 503 (2018).
Wang, S. C., Yu, C. F., Hong, J. H., Tsai, C. S. & Chiang, C. S. Radiation therapy-induced tumor invasiveness is associated with SDF-1-regulated macrophage mobilization and vasculogenesis. PLoS ONE 8, e69182 (2013).
Russell, J. S. & Brown, J. M. The irradiated tumor microenvironment: role of tumor-associated macrophages in vascular recovery. Front. Physiol. 4, 157 (2013).
Tabatabaei, P. et al. Radiotherapy induces an immediate inflammatory reaction in malignant glioma: a clinical microdialysis study. J. Neurooncol. 131, 83–92 (2017).
Bhat, K. P. L. et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 (2013).
Thomas, R. P. et al. Macrophage exclusion after radiation therapy (MERT): a first in human phase I/II trial using a CXCR4 inhibitor in glioblastoma. Clin. Cancer Res. 25, 6948–6957 (2019).
Akkari, L. et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl Med. 12, eaaw7843 (2020).
Poon, C. C. et al. Differential microglia and macrophage profiles in human IDH-mutant and -wild type glioblastoma. Oncotarget 10, 3129–3143 (2019).
van Dalen, F. J., van Stevendaal, M., Fennemann, F. L., Verdoes, M. & Ilina, O. Molecular repolarisation of tumour-associated macrophages. Molecules 24, 9 (2018).
Paolicelli, R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Chen, Y. & Xu, R. Drug repurposing for glioblastoma based on molecular subtypes. J. Biomed. Inf. 64, 131–138 (2016).
Jeanmougin, M. et al. Improved prognostication of glioblastoma beyond molecular subtyping by transcriptional profiling of the tumor microenvironment. Mol. Oncol. 14, 1016–1027 (2020).
Olar, A. & Aldape, K. D. Using the molecular classification of glioblastoma to inform personalized treatment. J. Pathol. 232, 165–177 (2014).
Sorensen, M. D., Dahlrot, R. H., Boldt, H. B., Hansen, S. & Kristensen, B. W. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 44, 185–206 (2018).
Zeiner, P. S. et al. Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain Pathol. 29, 513–529 (2019).
Villa, A., Della Torre, S. & Maggi, A. Sexual differentiation of microglia. Front. Neuroendocrinol. 52, 156–164 (2019).
Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Acknowledgements
The authors’ research is supported by grants from the Swedish Research Council (to B.J.), Swedish Cancer Foundation (to B.J.), Swedish Childhood Cancer Foundation (to K.B.), Swedish Cancer Society (to B.J.), Swedish Brain Foundation (to B.J.), Åke Wiberg Foundation (to M.C.), Petrus och Augusta Hedlund Foundation (to M.C.) and Karolinska Institutet Foundations (to B.J.).
Author information
Authors and Affiliations
Contributions
L.K. and B.J. contributed to all aspects of the article. M.C. and K.B. contributed substantially to discussions of the article content and participated in review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Neurology thanks B. Kaminska, R. Verhaak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Keane, L., Cheray, M., Blomgren, K. et al. Multifaceted microglia — key players in primary brain tumour heterogeneity. Nat Rev Neurol 17, 243–259 (2021). https://doi.org/10.1038/s41582-021-00463-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41582-021-00463-2
This article is cited by
-
Microglia-mediated drug substance transfer promotes chemoresistance in brain tumors: insights from an in vitro co-culture model using GCV/Tk prodrug system
Cancer Cell International (2024)
-
Artificial intelligence in neuro-oncology: advances and challenges in brain tumor diagnosis, prognosis, and precision treatment
npj Precision Oncology (2024)
-
Hypoxia drives shared and distinct transcriptomic changes in two invasive glioma stem cell lines
Scientific Reports (2024)
-
Modeling nervous system tumors with human stem cells and organoids
Cell Regeneration (2023)
-
CCL21-CCR7 signaling promotes microglia/macrophage recruitment and chemotherapy resistance in glioblastoma
Cellular and Molecular Life Sciences (2023)