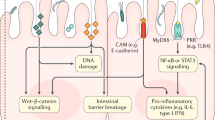
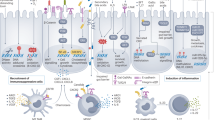
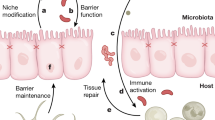
Abstract
Colorectal cancer (CRC) is a substantial source of global morbidity and mortality in dire need of improved prevention and treatment strategies. As our understanding of CRC grows, it is becoming increasingly evident that the gut microbiota, consisting of trillions of microorganisms in direct interface with the colon, plays a substantial role in CRC development and progression. Understanding the roles that individual microorganisms and complex microbial communities play in CRC pathogenesis, along with their attendant mechanisms, will help yield novel preventive and therapeutic interventions for CRC. In this Review, we discuss recent evidence concerning global perturbations of the gut microbiota in CRC, associations of specific microorganisms with CRC, the underlying mechanisms by which microorganisms potentially drive CRC development and the roles of complex microbial communities in CRC pathogenesis. While our understanding of the relationship between the microbiota and CRC has improved in recent years, our findings highlight substantial gaps in current research that need to be filled before this knowledge can be used to the benefit of patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
de Martel, C et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 13, 607–615 (2012).
Veettil, S. K. et al. Role of diet in colorectal cancer incidence. JAMA Netw. Open. 4, e2037341 (2021).
Clay, S. L., Fonseca-Pereira, D. & Garrett, W. S. Colorectal cancer: the facts in the case of the microbiota. J. Clin. Invest. 132, e155101 (2022).
Shaukat, A. & Levin, T. R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 19, 521–531 (2022).
Zhang, J. et al. Oral antibiotic use and risk of colorectal cancer in the United Kingdom, 1989–2012: a matched case–control study. Gut 68, 1971 (2019).
Lu, S. S. M. et al. Antibiotics use and subsequent risk of colorectal cancer: a Swedish nationwide population-based study. J. Natl Cancer Inst. 114, djab125 (2021).
Weng, L. et al. Antibiotics use and risk of colorectal neoplasia: an updated meta-analysis. Int. J. Colorectal Dis. 37, 2291–2301 (2022).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
He, Y. et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24, 1532–1535 (2018).
Keohane, D. M. et al. Microbiome and health implications for ethnic minorities after enforced lifestyle changes. Nat. Med. 26, 1089–1095 (2020).
Zhang, J. & Sears, C. L. Antibiotic use impacts colorectal cancer: a double-edged sword by tumor location? J. Natl Cancer Inst. 114, djab126 (2021).
Bartolomaeus, T. U. P. et al. Quantifying technical confounders in microbiome studies. Cardiovasc. Res. 117, 863–875 (2020).
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Liu, N.-N. et al. Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 7, 238–250 (2022).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019). Together with Thomas et al. (2019) and Liu et al. (2022), this study identified associations of microbial taxa and genes with CRC across multiple populations and study designs.
Mouradov, D. et al. Onco-microbial community profiling identifies clinico-molecular and prognostic subtypes of colorectal cancer. Gastroenterology 165, 104–120 (2023).
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Gao, R. et al. Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology 163, 1024–1037.e9 (2022).
Tjalsma, H., Boleij, A., Marchesi, J. R. & Dutilh, B. E. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582 (2012).
Zuo, W., Michail, S. & Sun, F. Metagenomic analyses of multiple gut datasets revealed the association of phage signatures in colorectal cancer. Front. Cell Infect. Microbiol. 12, 918010 (2022).
Hannigan, G. D., Duhaime, M. B., Ruffin, M. T., Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9, e02248-18 (2018).
Nakatsu, G. et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541.e5 (2018).
Chen, F. et al. Meta-analysis of fecal viromes demonstrates high diagnostic potential of the gut viral signatures for colorectal cancer and adenoma risk assessment. J. Adv. Res. 49, 103–114 (2022).
Coker, O. O. et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68, 654 (2019).
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020). This study profiled intratumoural microbiota across diverse human cancer types, revealing that each tumour type has a distinct microbial composition.
Niño, J. L. G. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022). This study showed that intratumoural microbiota localize to ‘microniches’ within tumours characterized by distinctive physiological and immunological states, suggesting that intratumoural microbiota affect the biology of the tumours.
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Klein, R. S. et al. Association of Streptococcus bovis with carcinoma of the colon. N. Engl. J. Med. 297, 800–802 (1977).
Pasquereau-Kotula, E., Martins, M., Aymeric, L. & Dramsi, S. Significance of Streptococcus gallolyticus subsp. gallolyticus association with colorectal cancer. Front. Microbiol. 9, 614 (2018).
Peek, R. M. & Crabtree, J. E. Helicobacter infection and gastric neoplasia. J. Pathol. 208, 233–248 (2006).
Knippel, R. J., Drewes, J. L. & Sears, C. L. The cancer microbiome: recent highlights and knowledge gaps. Cancer Discov. 11, 2378–2395 (2021).
Swidsinski, A. et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 115, 281–286 (1998).
Nougayrède, J.-P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Bossuet-Greif, N. et al. The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9, e02393-17 (2018).
Dziubańska-Kusibab, P. J. et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26, 1063–1069 (2020).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020). Together with Dziubańska-Kusibabh et al. (2020), this study identified a distinct mutational signature generated by colibactin in human CRC.
Iftekhar, A. et al. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 12, 1003 (2021).
Berger, H. & Meyer, T. F. Mechanistic dissection unmasks colibactin as a prevalent mutagenic driver of cancer. Cancer Cell 39, 1439–1441 (2021).
Tsunematsu, Y. et al. Mother-to-infant transmission of the carcinogenic colibactin-producing bacteria. BMC Microbiol. 21, 235 (2021).
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537 (2019).
Tang-Fichaux, M. et al. The polyphosphate kinase of Escherichia coli is required for full production of the genotoxin colibactin. mSphere 5, e01195-20 (2020).
Valguarnera, E. & Wardenburg, J. B. Good gone bad: one toxin away from disease for Bacteroides fragilis. J. Mol. Biol. 432, 765–785 (2019).
Sears, C. L. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin. Microbiol. Rev. 22, 349–369 (2009).
Boleij, A. et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 60, 208–215 (2015).
Wu, S., Lim, K.-C., Huang, J., Saidi, R. F. & Sears, C. L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl Acad. Sci. USA 95, 14979–14984 (1998).
Wu, S., Morin, P. J., Maouyo, D. & Sears, C. L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 12f4, 392–400 (2003).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009).
Rhee, K.-J. et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77, 1708–1718 (2009).
Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 203–214.e5 (2018). This study elucidated the immunological mechanism of enterotoxigenic B. fragilis-induced colon tumorigenesis in ApcMin mice.
Zhang, L. & Shay, J. W. Multiple roles of APC and its therapeutic implications in colorectal cancer.J. Natl Cancer Inst. 109, djw332 (2017).
Purcell, R. V. et al. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS ONE 12, e0171602 (2017).
Périchon, B. et al. Detection of Streptococcus gallolyticus and four other CRC-associated bacteria in patient stools reveals a potential “driver” role for enterotoxigenic Bacteroides fragilis. Front. Cell Infect. Microbiol. 12, 794391 (2022).
Zamani, S. et al. Enterotoxigenic Bacteroides fragilis: a possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front. Cell Infect. Microbiol. 9, 449 (2020).
Aitchison, A., Pearson, J. F., Purcell, R. V., Frizelle, F. A. & Keenan, J. I. Detection of Fusobacterium nucleatum DNA in primary care patient stool samples does not predict progression of colorectal neoplasia. PLoS ONE 17, e0269541 (2022).
Han, Y. W. Fusobacterium nucleatum: a commensal-turned pathogen. Curr. Opin. Microbiol. 23, 141–147 (2015).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012).
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-Cadherin/β-Catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Abed, J. et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20, 215–225 (2016).
Komiya, Y. et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 68, 1335 (2019).
Abed, J. et al. Colon cancer-associated Fusobacterium nucleatum may originate from the oral cavity and reach colon tumors via the circulatory system. Front. Cell Infect. Microbiol. 10, 400 (2020).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 (2017). This study showed that F. nucleatum increases chemoresistance of CRC cells by stimulating autophagy.
Queen, J. et al. Comparative analysis of colon cancer-derived Fusobacterium nucleatum subspecies: inflammation and colon tumorigenesis in murine models. mBio 13, e02991-21 (2022).
Brennan, C. A. et al. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes 13, 1987780 (2021).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β‐catenin modulator Annexin A1. EMBO Rep. 20, e47638 (2019).
Kumar, R. et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 13, e1006440 (2017).
Andres-Franch, M. et al. Streptococcus gallolyticus infection in colorectal cancer and association with biological and clinical factors. PLoS ONE 12, e0174305 (2017).
Aymeric, L. et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc. Natl Acad. Sci. USA 115, E283–E291 (2018).
Long, X. et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 4, 2319–2330 (2019).
Wang, X. et al. Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 inflammasome. Cancer Res. 81, 2745–2759 (2021).
Zhao, L. et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene 41, 4200–4210 (2022).
Nagaraja, V. & Eslick, G. D. Systematic review with meta‐analysis: the relationship between chronic Salmonella typhi carrier status and gall‐bladder cancer. Aliment. Pharm. Ther. 39, 745–750 (2014).
Scanu, T. et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17, 763–774 (2015).
Mughini-Gras, L. et al. Increased colon cancer risk after severe Salmonella infection. PLoS ONE 13, e0189721 (2018).
Duijster, J. W. et al. Association between Salmonella infection and colon cancer: a nationwide registry-based cohort study. Epidemiol. Infect. 149, e56 (2021).
Drewes, J. L. et al. Human colon cancer-derived Clostridioides difficile strains drive colonic tumorigenesis in mice. Cancer Discov. 12, 1873–1885 (2022).
He, Z. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68, 289–300 (2019).
Wu, N. et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 66, 462–470 (2013).
Okuda, S. et al. Profiling of host genetic alterations and intra-tumor microbiomes in colorectal cancer. Comput. Struct. Biotechnol. J. 19, 3330–3338 (2021).
Li, Y. et al. Gut microbiota signatures in tumor, para-cancerous, normal mucosa, and feces in colorectal cancer patients. Front. Cell Dev. Biol. 10, 916961 (2022).
Crobach, M. J. T. et al. Understanding Clostridium difficile colonization.Clin. Microbiol. Rev. 31, e00021-17 (2018).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Nguyen, L. H., Goel, A. & Chung, D. C. Pathways of colorectal carcinogenesis. Gastroenterology 158, 291–302 (2020).
Cao, Y. et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378, eabm3233 (2022).
Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019).
Xue, M. et al. Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685 (2019).
Xue, M., Wernke, K. M. & Herzon, S. B. Depurination of colibactin-derived interstrand cross-links. Biochemistry 59, 892–900 (2020).
Scuron, M. D., Boesze-Battaglia, K., Dlakić, M. & Shenker, B. J. The cytolethal distending toxin contributes to microbial virulence and disease pathogenesis by acting as a tri-perditious toxin. Front. Cell Infect. Microbiol. 6, 168 (2016).
Buc, E. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 8, e56964 (2013).
Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019).
Eaden, J. A., Abrams, K. R. & Mayberry, J. F. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 48, 526 (2001).
Raskov, H., Orhan, A., Christensen, J. P. & Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 124, 359–367 (2021).
Schmitt, M. & Greten, F. R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 21, 653–667 (2021).
Ellulu, M. S., Patimah, I., Khaza’ai, H., Rahmat, A. & Abed, Y. Obesity and inflammation: the linking mechanism and the complications. Arch. Med. Sci. 13, 851–863 (2017).
Tabung, F. K. et al. Association of dietary inflammatory potential with colorectal cancer risk in men and women. JAMA Oncol. 4, 366 (2018).
Geis, A. L. et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 5, 1098–1109 (2015).
Canli, Ö. et al. Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell 32, 869–883.e5 (2017).
Irrazabal, T. et al. Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat. Commun. 11, 1802 (2020).
Zea, A. H. et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 65, 3044–3048 (2005).
Huang, B. et al. Gr-1+ CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 66, 1123–1131 (2006).
Wang, K. et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 41, 1052–1063 (2014).
Gur, C. et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42, 344–355 (2015). This study showed that F. nucleatum inhibits antitumor immunity by binding its Fap2 adhesin to the TIGIT receptor expressed on natural killer cells and T cells.
Galaski, J. et al. Fusobacterium nucleatum CbpF mediates inhibition of T cell function through CEACAM1 activation. Front. Cell Infect. Microbiol. 11, 692544 (2021).
Peuker, K. et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immunity 55, 701–717.e7 (2022).
Zhang, X. et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe 31, 418–432.e8 (2023).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Kim, J. M. et al. Mitogen‐activated protein kinase and activator protein‐1 dependent signals are essential for Bacteroides fragilis enterotoxin‐induced enteritis. Eur. J. Immunol. 35, 2648–2657 (2005).
Wu, S. et al. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect. Immun. 72, 5832–5839 (2004).
Bao, Y. et al. Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 10, 675 (2019).
Cao, Y. et al. Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology 161, 1552–1566.e12 (2021).
Goodwin, A. C. et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA 108, 15354–15359 (2011).
Allen, J. et al. Colon tumors in enterotoxigenic Bacteroides fragilis (ETBF)-colonized mice do not display a unique mutational signature but instead possess host-dependent alterations in the APC gene. Microbiol. Spectr. 10, e01055-22 (2022).
Allen, J., Hao, S., Sears, C. L. & Timp, W. Epigenetic changes induced by Bacteroides fragilis toxin (BFT). Infect. Immun. 87, e00447-18 (2019).
Liu, Q.-Q. et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes 12, 1788900 (2020).
Meng, Q. et al. Fusobacterium nucleatum secretes amyloid‐like FadA to enhance pathogenicity. EMBO Rep. 22, e52891 (2021).
Zhang, Y. et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes 14, 2038852 (2022).
Lu, X. et al. Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. 40, 111127 (2022).
Chen, S. et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes 11, 511–521 (2020).
Hong, J. et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 70, 2123–2137 (2021).
Liu, J. et al. Proteomic characterization of outer membrane vesicles from gut mucosa-derived Fusobacterium nucleatum. J. Proteom. 195, 125–137 (2019).
Engevik, M. A. et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio 12, e02706-20 (2021).
Zakharzhevskaya, N. B. et al. Interaction of Bacteroides fragilis toxin with outer membrane vesicles reveals new mechanism of its secretion and delivery. Front. Cell Infect. Microbiol. 7, 2 (2017).
Vestby, L. K., Grønseth, T., Simm, R. & Nesse, L. L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 9, 59 (2020).
Dejea, C. M. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA 111, 18321–18326 (2014).
Drewes, J. L. et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34 (2017).
Johnson, C. H. et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21, 891–897 (2015).
Tomkovich, S. et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest. 129, 1699–1712 (2019).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Baran, B. et al. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterol. Res. 11, 264–273 (2018).
Mangone, L. et al. Colon cancer survival differs from right side to left side and lymph node harvest number matter. BMC Public Health 21, 906 (2021).
Cai, J., Sun, L. & Gonzalez, F. J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 30, 289–300 (2022).
Ajouz, H., Mukherji, D. & Shamseddine, A. Secondary bile acids: an underrecognized cause of colon cancer. World J. Surg. Oncol. 12, 164 (2014).
Fu, T. et al. FXR regulates intestinal cancer stem cell proliferation. Cell 176, 1098–1112.e18 (2019).
Sorrentino, G. et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 159, 956–968.e8 (2020).
Booth, L. A., Gilmore, I. T. & Bilton, R. F. Secondary bile acid induced DNA damage in HT29 cells: are free radicals involved? Free Radic. Res. 26, 135–144 (2009).
Zhang, H., Xu, H., Zhang, C., Tang, Q. & Bi, F. Ursodeoxycholic acid suppresses the malignant progression of colorectal cancer through TGR5-YAP axis. Cell Death Discov. 7, 207 (2021).
Alberts, D. S. et al. Phase III trial of ursodeoxycholic acid to prevent colorectal adenoma recurrence. J. Natl Cancer Inst. 97, 846–853 (2005).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020).
Li, W. et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377.e9 (2021).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020). This study determined that microbial secondary bile acid production regulates regulatory T cell differentiation in the gut.
Sato, Y. et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 599, 458–464 (2021).
Paik, D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022). This study identified the bacterial species and enzymes that produce two prominent secondary bile acids, 3-oxolithocholic acid and isolithocholic acid, which inhibit TH17 cell differentiation in the gut.
Lavoie, S. et al. Expression of free fatty acid receptor 2 by dendritic cells prevents their expression of interleukin 27 and is required for maintenance of mucosal barrier and immune response against colorectal tumors in mice. Gastroenterology 158, 1359–1372.e9 (2020).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Donohoe, D. R. et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 4, 1387–1397 (2014).
Wang, T. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329 (2012).
O’Keefe, S. J. D. et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 6, 6342 (2015).
Chen, D. et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467 (2020). This study showed that administration of Clostridium butyricum could reduce high-fat diet-induced colorectal tumorigenesis in ApcMin mice.
Deehan, E. C. et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe 27, 389–404.e6 (2020).
Belcheva, A. et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell 158, 288–299 (2014).
Okumura, S. et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 12, 5674 (2021). Together with Belcheva et al. (2014), this study demonstrated that certain bacteria in the gut can potentiate colorectal tumorigenesis through butyrate production, highlighting the context-dependent and incompletely understood role of butyrate in CRC development.
Bultman, S. J. & Jobin, C. Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe 16, 143–145 (2014).
Donohoe, D. R. et al. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 48, 612–626 (2012).
Sinicrope, F. A. Increasing incidence of early-onset colorectal cancer. N. Engl. J. Med. 386, 1547–1558 (2022).
Han, J.-X. et al. Microbiota-derived tryptophan catabolites mediate the chemopreventive effects of statins on colorectal cancer. Nat. Microbiol. 8, 919–933 (2023).
Bell, H. N. et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 40, 185–200.e6 (2022).
Montalban-Arques, A. et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe 29, 1573–1588.e7 (2021).
Ryu, S. W. et al. Gut microbiota Eubacterium callanderi exerts anti-colorectal cancer activity. Microbiol. Spectr. 10, e02531-22 (2022).
Xu, H., Luo, H., Zhang, J., Li, K. & Lee, M.-H. Therapeutic potential of Clostridium butyricum anticancer effects in colorectal cancer. Gut Microbes 15, 2186114 (2023).
Dikeocha, I. J., Al-Kabsi, A. M., Chiu, H.-T. & Alshawsh, M. A. Faecalibacterium prausnitzii ameliorates colorectal tumorigenesis and suppresses proliferation of HCT116 colorectal cancer cells. Biomedicines 10, 1128 (2022).
Kim, O. Y. et al. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 8, 626 (2017).
Qing, S. et al. Biomineralized bacterial outer membrane vesicles potentiate safe and efficient tumor microenvironment reprogramming for anticancer therapy. Adv. Mater. 32, 2002085 (2020).
Zheng, D.-W. et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728 (2019).
Jia, F. et al. Optimized antimicrobial peptide jelleine-I derivative Br-J-I inhibits Fusobacterium nucleatum to suppress colorectal cancer progression. Int. J. Mol. Sci. 24, 1469 (2023).
Holt, R. A. Oncomicrobial vaccines: the potential for a Fusobacterium nucleatum vaccine to improve colorectal cancer outcomes. Cell Host Microbe 31, 141–145 (2023).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602 (2021). Together with Baruch et al. (2021), this study demonstrated the efficacy of faecal microbiota transplant therapy in overcoming immunotherapy resistance in melanoma, serving as a goal for future microbiome-directed therapeutics in CRC.
Walter, J. & Shanahan, F. Fecal microbiota-based treatment for recurrent Clostridioides difficile infection. Cell 186, 1087 (2023).
Mullard, A. FDA approves second microbiome-based C. difficile therapy. Nat. Rev. Drug Discov. 22, 436 (2023).
Tomkovich, S. et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 77, 2620–2632 (2017).
Hwang, S. et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int. J. Med. Sci. 17, 145–152 (2020).
Xi, Y. & Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 14, 101174 (2021).
Jasperson, K. W., Tuohy, T. M., Neklason, D. W. & Burt, R. W. Hereditary and familial colon cancer. Gastroenterology 138, 2044–2058 (2010).
Jiao, S. et al. Estimating the heritability of colorectal cancer. Hum. Mol. Genet. 23, 3898–3905 (2014).
Hossain, M. S. et al. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers 14, 1732 (2022).
Helsingen, L. M. & Kalager, M. Colorectal cancer screening — approach, evidence, and future directions. NEJM Evid. https://doi.org/10.1056/EVIDra2100035 (2022).
Benson, A. B. et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl Compr. Canc. Netw. 19, 329–359 (2021).
Ooki, A., Shinozaki, E. & Yamaguchi, K. Immunotherapy in colorectal cancer: current and future strategies. J. Anus Rectum Colon. 5, 11–24 (2021).
Dow, L. E. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015).
Acknowledgements
This work was funded by the US National Institutes of Health (R01CA196845 and U2CCA233291), Cancer Research UK (Cancer Grand Challenges OPTIMISTICC team grant A27140) and Janssen. The authors are grateful for the contributions of members of the Sears Laboratory over time.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to the design of the manuscript. M.T.W. wrote most of the manuscript and created all display items. C.L.S. wrote the Conclusions and clinical applications section and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Sears Laboratory receives research support through Johns Hopkins University School of Medicine from Janssen and Bristol Myers Squibb.
Peer review
Peer review information
Nature Reviews Microbiology thanks Andrew Chan, Subbaya Subramanian, Yiqing Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
White, M.T., Sears, C.L. The microbial landscape of colorectal cancer. Nat Rev Microbiol 22, 240–254 (2024). https://doi.org/10.1038/s41579-023-00973-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-023-00973-4
This article is cited by
-
Whittling down the bacterial subspecies that might drive colon cancer
Nature (2024)
-
Association of Fusobacterium nucleatum infection with the clinicopathological characteristics in colorectal cancer patients
Molecular Biology Reports (2024)