Abstract
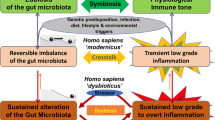
Symbiotic microorganisms inhabiting the gastrointestinal tract promote health by decreasing susceptibility to infection and enhancing resistance to a range of diseases. In this Review, we discuss our increasing understanding of the impact of the microbiome on the mammalian host and recent efforts to culture and characterize intestinal symbiotic microorganisms that produce or modify metabolites that impact disease pathology. Manipulation of the intestinal microbiome has great potential to reduce the incidence and/or severity of a wide range of human conditions and diseases, and the biomedical research community now faces the challenge of translating our understanding of the microbiome into beneficial medical therapies. Our increasing understanding of symbiotic microbial species and the application of ecological principles and machine learning are providing exciting opportunities for microbiome-based therapeutics to progress from faecal microbiota transplantation to the administration of precisely defined and clinically validated symbiotic microbial consortia that optimize disease resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4554–4561 (2011).
Borbet, T. C., Zhang, X., Muller, A. & Blaser, M. J. The role of the changing human microbiome in the asthma pandemic. J. Allergy Clin. Immunol. 144, 1457–1466 (2019).
Becattini, S., Taur, Y. & Pamer, E. G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478 (2016).
Gottfried, J. History Repeating? Avoiding a Return to the Pre-antibiotic Age. Harvard Library http://nrs.harvard.edu/urn-3:HUL.InstRepos:8889467 (2005).
Peled, J. U. et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N. Engl. J. Med. 382, 822–834 (2020). This large, multi-institutional clinical study spanning three continents demonstrates the association of microbiota diversity with survival following haematopoietic cell transplantation.
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Fischbach, M. A. Microbiome: focus on causation and mechanism. Cell 174, 785–790 (2018).
Pamer, E. G. Fecal microbiota transplantation: effectiveness, complexities, and lingering concerns. Mucosal Immunol. 7, 210–214 (2014).
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Goodman, A. L. et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011).
Fehlner-Peach, H. et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 26, 680–690 (2019).
Sorbara, M. T. et al. Functional and genomic variation between human-derived isolates of Lachnospiraceae reveals inter- and intra-species diversity. Cell Host Microbe 28, 134–146 (2020). This study demonstrates genomic differences between symbiotic bacterial strains, many of which may impact their potential as live biotherapeutic products.
Guo, C. J. et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science https://doi.org/10.1126/science.aav1282 (2019). A pioneering study demonstrating the impact of multiple genes on the production of metabolites by C. sporogenes.
Funabashi, M. et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020).
Danne, C., Rolhion, N. & Sokol, H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat. Rev. Gastroenterol. Hepatol. 18, 503–513 (2021).
Abt, M. C., McKenney, P. T. & Pamer, E. G. Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 14, 609–620 (2016).
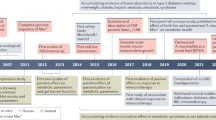
Eiseman, B., Silen, W., Bascom, G. S. & Kauvar, A. J. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44, 854–859 (1958).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Kassam, Z., Lee, C. H., Yuan, Y. & Hunt, R. H. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol. 108, 500–508 (2013).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Smillie, C. S. et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240 (2018).
Kakihana, K. et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 128, 2083–2088 (2016).
van Lier, Y. F. et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aaz8926 (2020).
Zhao, Y. et al. Safety and efficacy of fecal microbiota transplantation for grade IV steroid refractory GI-GvHD patients: interim results from FMT2017002 trial. Front. Immunol. 12, 678476 (2021).
Kootte, R. S. et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 26, 611–619 (2017).
de Groot, P. et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut 70, 92–105 (2021).
Yu, E. W. et al. Fecal microbiota transplantation for the improvement of metabolism in obesity: the FMT-TRIM double-blind placebo-controlled pilot trial. PLoS Med. 17, e1003051 (2020).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A. & Alm, E. J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1784 (2017).
Rossen, N. G. et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118 (2015).
Moayyedi, P. et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109 (2015).
Paramsothy, S. et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017).
Costello, S. P. et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 321, 156–164 (2019).
Sokol, H. et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: a pilot randomized controlled study. Microbiome 8, 12 (2020).
Kong, L. et al. Linking strain engraftment in fecal microbiota transplantation with maintenance of remission in Crohn’s disease. Gastroenterology 159, 2193–2202 (2020).
Witjes, J. J. et al. Donor fecal microbiota transplantation alters gut microbiota and metabolites in obese individuals with steatohepatitis. Hepatol. Commun. 4, 1578–1590 (2020).
DeFilipp, Z., Bloom, P. P., Hohmann, E. L. & Group, F. M. T. S. Drug-resistant bacteremia after fecal microbiota transplant. N. Engl. J. Med. 382, 1961–1962 (2020).
Terveer, E. M. et al. Human transmission of blastocystis by fecal microbiota transplantation without development of gastrointestinal symptoms in recipients. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciz1122 (2019).
Gareau, M. G., Sherman, P. M. & Walker, W. A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514 (2010).
Kim, Y. G. et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319 (2017). This is an important study demonstrating the important role of the order Clostridiales in providing colonization resistance against a range of enteric pathogens.
Kim, S. G. et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572, 665–669 (2019). This study demonstrates that lantibiotic production by an obligate anaerobic commensal bacterium can markedly reduce intestinal colonization by vancomycin-resistant E. faecium.
Becattini, S. et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J. Exp. Med. 214, 1973–1989 (2017).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Byndloss, M. X. et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017).
Rivera-Chavez, F. et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). This landmark study introduces the rational development of a commensal consortium to drive a beneficial interaction with the host.
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Feehley, T. et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 25, 448–453 (2019). This study demonstrates that butyrate-producing commensal bacteria, such as A. caccae, can provide protection against food allergies.
Stefka, A. T. et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl Acad. Sci. USA 111, 13145–13150 (2014).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020). This study shows that modification of secondary bile acids by resident commensal bacteria of the lower intestinal tract can impact differentiation of T cells by impacting dendritic cell function.
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Kamdar, K. et al. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe 19, 21–31 (2016).
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Hall, A. B. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017).
Laverde Gomez, J. A. et al. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Envron. Microbiol. 21, 259–271 (2019).
Rey, F. E. et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285, 22082–22090 (2010).
Koenig, J. E. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4578–4585 (2011).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Wexler, A. G. & Goodman, A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017).
Sato, Y. et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature https://doi.org/10.1038/s41586-021-03832-5 (2021). This study demonstrates that a wider range of commensal bacteria can modify secondary bile acids, leading to the production of isoall lithocholic acid, which has high activity against intestinal pathogens, such as C. difficile.
Li, W. et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377 (2021). This study demonstrates a mechanism by which secondary bile acids can enhance the differentiation of Treg cells.
Porter, N. T. & Martens, E. C. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu. Rev. Microbiol. 71, 349–369 (2017).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018).
Russell, A. B. et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 (2014).
Ross, B. D. et al. Human gut bacteria contain acquired interbacterial defence systems. Nature 575, 224–228 (2019).
Coyne, M. J. et al. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat. Commun. 10, 3460 (2019). This study demonstrates one mechanism by which members of the order Bacteoridales can compete with the complex microbiota of the lower intestinal tract.
Shumaker, A. M., Laclare McEneany, V., Coyne, M. J., Silver, P. A. & Comstock, L. E. Identification of a fifth antibacterial toxin produced by a single bacteroides fragilis strain. J. Bacteriol. https://doi.org/10.1128/JB.00577-18 (2019).
Garcia-Bayona, L. & Comstock, L. E. Bacterial antagonism in host-associated microbial communities. Science https://doi.org/10.1126/science.aat2456 (2018).
Chia, L. W. et al. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van. Leeuwenhoek 111, 859–873 (2018).
Caesar, R., Tremaroli, V., Kovatcheva-Datchary, P., Cani, P. D. & Backhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22, 658–668 (2015).
Ribo, S. et al. Increasing breast milk betaine modulates Akkermansia abundance in mammalian neonates and improves long-term metabolic health. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abb0322 (2021).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Seregin, S. S. et al. NLRP6 protects Il10-/- mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 19, 733–745 (2017).
Cirstea, M., Radisavljevic, N. & Finlay, B. B. Good bug, bad bug: breaking through microbial stereotypes. Cell Host Microbe 23, 10–13 (2018).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Katoh, T. et al. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms https://doi.org/10.3390/microorganisms8040481 (2020).
Sugahara, H. et al. Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci. Rep. 5, 13548 (2015).
Sun, S. et al. Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc. Natl Acad. Sci. USA 117, 27509–27515 (2020).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Xiao, J. et al. Probiotic Bifidobacterium breve in improving cognitive functions of older adults with suspected mild cognitive impairment: a randomized, double-blind, placebo-controlled trial. J. Alzheimers Dis. 77, 139–147 (2020).
Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 (2015).
Zeng, M. Y., Inohara, N. & Nunez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26 (2017).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007).
Sorbara, M. T. et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J. Exp. Med. 216, 84–98 (2019).
Jacobson, A. et al. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24, 296–307 (2018).
Osbelt, L. et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog. 16, e1008448 (2020).
Stoma, I. et al. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa068 (2020).
Taur, Y. et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914 (2012).
Wassenaar, T. M. Insights from 100 years of research with probiotic E. coli. Eur. J. Microbiol. Immunol. 6, 147–161 (2016).
Gurry, T. et al. Predictability and persistence of prebiotic dietary supplementation in a healthy human cohort. Sci. Rep. 8, 12699 (2018).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Ang, Q. Y. et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell 181, 1263–1275 (2020).
von Schwartzenberg, R. J. et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 595, 272–277 (2021). This study demonstrates that severe caloric restriction can markedly alter microbiota composition and may provide advantages to enteric pathogens.
Korem, T. et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 25, 1243–1253 (2017).
Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 22, 971–982 (2015).
Carmody, R. N. et al. Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 4, 2052–2063 (2019).
Gehrig, J. L. et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science https://doi.org/10.1126/science.aau4732 (2019).
Chen, R. Y. et al. A microbiota-directed food intervention for undernourished children. N. Engl. J. Med. 384, 1517–1528 (2021).
Delannoy-Bruno, O. et al. Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature 595, 91–95 (2021). This article drives home the impact of ingested fibre on microbiota composition.
Galvez, E. J. C. et al. Distinct polysaccharide utilization determines interspecies competition between intestinal Prevotella spp. Cell Host Microbe 28, 838–852 (2020).
Patnode, M. L. et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 179, 59–73 (2019).
Pamer, E. G. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538 (2016).
Bohnhoff, M. & Miller, C. P. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J. Infect. Dis. 111, 117–127 (1962).
Freter, R. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. II. The inhibitory mechanism. J. Infect. Dis. 110, 38–46 (1962).
Schaedler, R. W., Dubs, R. & Costello, R. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122, 77–82 (1965).
Wymore Brand, M. et al. The altered Schaedler flora: continued applications of a defined murine microbial community. ILAR J. 56, 169–178 (2015).
Kane, M. et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334, 245–249 (2011).
Stecher, B. et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6, e1000711 (2010).
van der Waaij, D., Vossen, J. M., Altes, C. K. & Hartgrink, C. Reconventionalization following antibiotic decontamination in man and animals. Am. J. Clin. Nutr. 30, 1887–1895 (1977).
Tvede, M. & Rask-Madsen, J. Bacteriotherapy for Clostridium difficile diarrhoea. Lancet 335, 110 (1990).
Lawley, T. D. et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995 (2012).
Caballero, S. et al. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21, 592–602 (2017).
Walker, M. C. et al. Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genomics 21, 387 (2020).
Brugiroux, S. et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215 (2016).
Eberl, C. et al. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe 29, 1680–1692 (2021).
Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016).
Nagao-Kitamoto, H. et al. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 26, 608–617 (2020).
Caballero-Flores, G., Pickard, J. M., Fukuda, S., Inohara, N. & Nunez, G. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28, 526–533 (2020).
Faith, J. J., Ahern, P. P., Ridaura, V. K., Cheng, J. & Gordon, J. I. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra211 (2014).
Atarashi, K. et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365 (2017).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Poyet, M. et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019).
Forster, S. C. et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat. Biotechnol. 37, 186–192 (2019).
Zou, Y. et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 37, 179–185 (2019).
Bisanz, J. E. et al. A genomic toolkit for the mechanistic dissection of intractable human gut bacteria. Cell Host Microbe 27, 1001–1013 (2020).
Venturelli, O. S. et al. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, e8157 (2018). This study begins to dissect the interactions between members of an assembled consortium of commensal bacterial strains using computational platforms.
Stein, R. R. et al. Computer-guided design of optimal microbial consortia for immune system modulation. eLife https://doi.org/10.7554/eLife.30916 (2018).
Mimee, M., Tucker, A. C., Voigt, C. A. & Lu, T. K. Programming a human commensal bacterium, bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71 (2015).
Lim, B., Zimmermann, M., Barry, N. A. & Goodman, A. L. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169, 547–558 (2017).
Garcia-Bayona, L. & Comstock, L. E. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio https://doi.org/10.1128/mBio.01762-19 (2019).
Bhattarai, Y. et al. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 23, 775–785 (2018).
Bhattarai, Y. et al. Bacterially derived tryptamine increases mucus release by activating a host receptor in a mouse model of inflammatory bowel disease. iScience 23, 101798 (2020).
Shepherd, E. S., DeLoache, W. C., Pruss, K. M., Whitaker, W. R. & Sonnenburg, J. L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438 (2018).
Quinn, R. A. et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129 (2020).
Guo, C. J. et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 168, 517–526 (2017).
Heilbronner, S., Krismer, B., Brotz-Oesterhelt, H. & Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-021-00569-w (2021).
Chen, H. et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 177, 1217–1231 (2019).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Env. Microbiol. 19, 29–41 (2017).
Reichardt, N. et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335 (2014).
Donohoe, D. R. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 (2011).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Haak, B. W. et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 131, 2978–2986 (2018).
Alavi, S. et al. Interpersonal gut microbiome variation drives susceptibility and resistance to cholera infection. Cell 181, 1533–1546 (2020).
Ridlon, J. M., Kang, D. J. & Hylemon, P. B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006).
Sorg, J. A. & Sonenshein, A. L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512 (2008).
Devlin, A. S. & Fischbach, M. A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 (2015).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020). This study demonstrates that secondary bile acid modifications can impact the differentiation of pro-inflammatory TH17 cells.
Sherkow, J. S. & Greely, H. T. The history of patenting genetic material. Annu. Rev. Genet. 49, 161–182 (2015).
FDA. Early Clinical Trials With Live Biotherapeutic Products: Chemistry, Manufacturing, and Control Information. FDA https://www.fda.gov/regulatory-information/search-fda-guidance-documents/early-clinical-trials-live-biotherapeutic-products-chemistry-manufacturing-and-control-information (2016).
European Pharmacopoeia Commission. 3053E general monograph on live biotherapeutic products Vol. 9 (European Pharmacopoeia Commission, 2019).
Paquet, J. C. et al. Entering first-in-human clinical study with a single-strain live biotherapeutic product: input and feedback gained from the EMA and the FDA. Front. Med. 8, 716266 (2021).
Blount, K. F., Shannon, W. D., Deych, E. & Jones, C. Restoration of bacterial microbiome composition and diversity among treatment responders in a phase 2 trial of RBX2660: an investigational microbiome restoration therapeutic. Open Forum Infect. Dis. 6, ofz095 (2019).
Dubberke, E. R. et al. Results from a randomized, placebo-controlled clinical trial of a RBX2660-a microbiota-based drug for the prevention of recurrent Clostridium difficile infection. Clin. Infect. Dis. 67, 1198–1204 (2018).
Orenstein, R. et al. Safety and durability of RBX2660 (microbiota suspension) for recurrent Clostridium difficile infection: results of the PUNCH CD study. Clin. Infect. Dis. 62, 596–602 (2016).
Dubberke, E. R. et al. Clearance of vancomycin-resistant Enterococcus concomitant with administration of a microbiota-based drug targeted at recurrent Clostridium difficile infection. Open. Forum Infect. Dis. https://doi.org/10.1093/ofid/ofw133 (2016).
Langdon, A. et al. Microbiota restoration reduces antibiotic-resistant bacteria gut colonization in patients with recurrent Clostridioides difficile infection from the open-label PUNCH CD study. Genome Med. 13, 28 (2021).
Khanna, S. et al. RBX7455, a room temperature-stable, orally-administered investigational live biotherapeutic, is safe, effective, and shifts patients’ microbiomes in a phase 1 study for recurrent Clostridioides difficile infections. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1430 (2020).
Kao, D. et al. The effect of a microbial ecosystem therapeutic (MET-2) on recurrent Clostridioides difficile infection: a phase 1, open-label, single-group trial. Lancet Gastroenterol. Hepatol. 6, 282–291 (2021).
Bobilev, D. et al. 1953. VE303, a rationally designed bacterial consortium for prevention of recurrent Clostridioides difficile (C. difficile) infection (rCDI), stably restores the gut microbiota after vancomycin (vanco)-induced dysbiosis in adult healthy volunteers (HV). Open. Forum Infect. Dis. 6, S60 (2019).
Acknowledgements
This work was supported by US NIH grants R01 AI095706, P01 CA023766, U01 AI124275, and R01 AI042135 to E.G.P. and the Duchossois Family Institute of the University of Chicago. M.T.S. was supported by a postdoctoral fellowship from the Canadian Institute for Health Research (FRN#152527) and a postdoctoral fellowship from Emerald Foundation Inc.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
E.G.P. serves on the advisory board of Diversigen, has received speaker honoraria from Bristol Myers Squibb, Celgene, Seres Therapeutics, MedImmune, Novartis and Ferring Pharmaceuticals, is an inventor on patent applications WPO2015179437A1, entitled “Methods and compositions for reducing Clostridium difficile infection”, and WO2017091753A1, entitled “Methods and compositions for reducing vancomycin-resistant enterococci infection or colonization”, and holds patents that receive royalties from Seres Therapeutics Inc. M.T.S. declares no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks J. Faith and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Microbiota restoration
-
Re-establishment of the diversity, density and beneficial functions of the microbiota following perturbation.
- Colonization resistance
-
Inhibition of exogenous species, including pathogens, by the resident microbiota.
- Colitis
-
Inflammation occurring in the large intestine.
- Engraftment
-
Establishment of a replicating, self-sustaining population of an introduced species or strain in a community.
- Alpha diversity
-
Measure of community diversity within a sample based on taxonomic richness (number of species) and evenness.
- Hepatic steatosis
-
A condition of non-alcoholic fatty liver disease defined by the accumulation of intrahepatic fat to greater than 5% of liver weight.
- Bacteroicins
-
Short ribosomally synthesized antimicrobial peptides produced by bacteria that often target closely related species but are, in general, not harmful to producing bacteria owing to immune mechanisms.
- Mucins
-
A family of large, highly glycosylated proteins secreted at mucosal surfaces. In the intestine, secreted mucins form a protective and selective barrier between epithelial cells and luminal contents.
- Checkpoint immunotherapy
-
Cancer therapy that functions by promoting an antitumour immune response by targeting proteins such as CTLA4 or PD1/PDL1 that normally function to limit immune responses.
Rights and permissions
About this article
Cite this article
Sorbara, M.T., Pamer, E.G. Microbiome-based therapeutics. Nat Rev Microbiol 20, 365–380 (2022). https://doi.org/10.1038/s41579-021-00667-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-021-00667-9
This article is cited by
-
Case report: Aberrant fecal microbiota composition of an infant diagnosed with prolonged intestinal botulism
Gut Pathogens (2024)
-
Short-chain fatty acid-producing bacterial strains attenuate experimental ulcerative colitis by promoting M2 macrophage polarization via JAK/STAT3/FOXO3 axis inactivation
Journal of Translational Medicine (2024)
-
Therapeutics for neurodegenerative diseases by targeting the gut microbiome: from bench to bedside
Translational Neurodegeneration (2024)
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)
-
Exploiting gut microbial traits and trade-offs in microbiome-based therapeutics
Nature Reviews Bioengineering (2024)