Abstract
It is now well known that non-coding RNAs (ncRNAs), rather than protein-coding transcripts, are the preponderant RNA transcripts. NcRNAs, particularly microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), are widely appreciated as pervasive regulators of multiple cancer hallmarks such as proliferation, apoptosis, invasion, metastasis, and genomic instability. Despite recent discoveries in cancer therapy, resistance to chemotherapy, radiotherapy, targeted therapy, and immunotherapy continue to be a major setback. Recent studies have shown that ncRNAs also play a major role in resistance to different cancer therapies by rewiring essential signaling pathways. In this review, we present the intricate mechanisms through which dysregulated ncRNAs control resistance to the four major types of cancer therapies. We will focus on the current clinical implications of ncRNAs as biomarkers to predict treatment response (intrinsic resistance) and to detect resistance to therapy after the start of treatment (acquired resistance). Furthermore, we will present the potential of targeting ncRNA to overcome cancer treatment resistance, and we will discuss the challenges of ncRNA-targeted therapy—especially the development of delivery systems.
Similar content being viewed by others
Introduction - overview of ncRNAs
The cataloging of the non-coding RNA (ncRNA) world is constantly and dramatically changing. Recent findings underscore the fact that a number of ncRNA transcripts can code for micropeptides (of less than 100 amino acids) that play functional roles in normal and pathological processes, including cancer.1 These novel data show that at least some ncRNAs have either both functional coding and non-coding capabilities or are, in fact, coding transcripts for non-classic peptides. Hence, we are facing the question of what ncRNA means.
Currently, the most studied types of “classic ncRNAs” are microRNAs (miRNAs),2 long-non-coding RNAs (lncRNAs),3 and circular RNAs (circRNAs).4 MiRNAs are short RNAs that originate from longer stem-loop structures and can bind and inhibit mRNAs.5 The biogenesis of miRNAs is a multistep process. MiRNAs are transcribed as primary miRNAs (pri-miRNAs) and processed in the nucleus by Drosha and Dgcr8 into precursor miRNAs (pre-miRNAs). After they are exported to the cytoplasm, pre-miRNAs are cleaved to form an miRNA/miRNA duplex. Only one of the two miRNAs formed will exert its inhibitory function, the other one being degraded.6 Several other unconventional miRNA functions have been reported, including binding and inhibiting proteins, activating Toll-like receptors, coding for peptides, activating the translation of mRNAs, inhibiting mitochondrial transcripts, triggering transcription, and inhibiting nuclear ncRNAs,7 making miRNAs complex and versatile molecules (Fig. 1a). The total number of known human miRNAs is in continuous expansion and currently includes 1917 precursors and 2654 mature molecules (miRBase, release 22.1).8 Many additional miRNAs, mostly with tissue-specific distribution, have also been discovered.9 Most of these genes are conserved between species.10
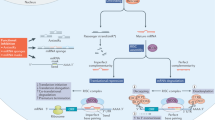
Biogenesis and function of miRNAs, lncRNAs and circRNAs. a MIRNAs are transcribed as primary miRNAs (pri-miRNAs) that contain the characteristic stem-loop structure. Pri-miRNAs are processed in the nucleus by Drosha and DGCR8 and transformed into precursor miRNAs (pre-miRNAs). Pre-miRNAs are transported from the nucleus to the cytoplasm via Exportin 5 and then are turned into an miRNA duplex after being cleaved by Dicer. One strand of the miRNA duplex is incorporated as part of the miRNA-induced silencing complex (RISC), and the second strand is degraded. By base-pairings between miRNAs and their target mRNA, the RISC binds an mRNA and suppresses its translation or induces its degradation. Additionally, there are unconventional/atypical miRNA functions such as activating Toll-like receptors (TLRs), binding non-AGO proteins, binding other ncRNAs (sponging), and regulating transcription. b Most lncRNAs have a biogenesis similar to mRNAs (although several exceptions exist), being capped, spliced, and adenylated. The mature lncRNAs adopt complex 3D structures that give them their multivalent functions. The function of lncRNAs can be divided according to their cellular localization: bound to chromatin (often cis functions), intranuclear (usually trans functions), and intracytoplasmic (trans functions). LncRNAs bound to chromatin usually function as regulators of transcription and induce chromosome looping and histone modifications. Nuclear lncRNAs can assemble paraspeckles and interact with nuclear proteins. Cytoplasmic lncRNAs bind mRNAs and act as decoys, guides, and scaffolds to transcriptionally or post-transcriptionally regulate downstream target genes, bind proteins to modify their function and stability, code for micropeptides that are being translated, and bind other ncRNA species (including miRNAs). c CircRNAs have multiple biogenesis mechanisms, but a common event for all is back-splicing. Back-splicing can be induced by protein dimerization, sequence complementarity of flanking introns, exon skipping mechanisms, and intron lariat debranching. After forming an uninterrupted RNA loop, the transcript is exported into the cytoplasm, where it serves as an miRNA sponge that inhibits miRNAs to regulate the expression of target genes, as a decoy of RNA-binding proteins to modulate gene expression or translation, or as a platform for protein-protein interaction; additionally, these transcripts also can be translated into micropeptides. As observed, there is direct crosstalk between lncRNAs and miRNAs and between circRNAs and miRNAs via sponging, creating a network of ncRNA molecules.
MiRNAs’ role in cancer was revealed in 2002, when it was discovered that in chronic lymphocytic leukemia (CLL) MIR15 and MIR16 are frequently deleted and their transcripts downregulated.11 Currently, miRNAs have been reported to be dysregulated in each type of investigated cancer.12,13,14 The complexity of miRNAs’ mechanisms of action and the multitude of targets has made it difficult for researchers to translate their findings into clinical practice,15 and a better understanding of their role in oncology is necessary.
LncRNAs are by far the most complex type of ncRNAs, being arbitrarily defined as RNA molecules over 200 nucleotides long that are usually not translated into proteins. This definition is unfortunately vague. For example, because of their length, several primary miRNAs are considered to be lncRNAs if they have a function on their own, and of all ncRNAs, lncRNAs have the highest potential of coding peptides (this has been confirmed several times).1 The biogenesis of lncRNAs is similar to that of mRNAs, many of them being spliced, capped, and poly-adenylated. The complexity of these transcripts comes from their multifaceted 3D structure, which rapidly changes and gives them the ability to perform multiple functions.16,17 LncRNAs have cis (performed in the proximity of their transcription site) and trans (performed distant from the transcription site) functions.18 Typical cis functions are related to DNA transcription, chromatin modifications, and chromosomal looping. Well-defined trans functions include binding to mRNAs and changing their stability, binding to proteins and altering their function, interacting with other ncRNAs, and facilitating the assembly of paraspeckles19,20 (Fig. 1b). The research on lncRNA surged after the 2003 discovery of MALAT1’s involvement in the metastasis of non-small cell lung cancer (NSCLC).21 Similar to miRNAs, lncRNAs’ role in cancer is now well studied but only rarely translated into clinical practice, the exception being PCA3 as a biomarker for prostate cancer.22
A complementary approach is to classify lncRNA from a phylogenetic standpoint. LncRNAs can be transcripts of ultraconserved elements that are identical in mice, rats, and humans.23 These lncRNAs are named transcribed-ultraconserved regions and because of their high degree of conservation are expected to have essential functions.24 On the other hand, are the more recently emerged transcripts, the primate-specific lncRNAs, which often contain transcribed pyknons in their structure.25,26 Pyknons are short, primate-specific, repetitive DNA motifs that are often localized in DNA-fragile sites and are transcribed as part of lncRNAs.25,27 These lncRNAs containing pyknons have low expression levels in normal cells; however, their expression level spikes in malignant and immune cells, making them ideal candidates for future therapies.20,28
CircRNAs, the third major class of ncRNAs, are characterized by their specific structure. CircRNAs are covalently closed uninterrupted loops, where the 3’ and 5’ ends are joined together.29 Because of this structure, circRNAs are more stable than other RNA types.29 CircRNAs have a complex and multifaceted biogenesis, for which multiple mechanisms have been described over the past years. CircRNAs can be generated by exon skipping mechanisms, intron lariat debranching, intron pairing, and RNA binding proteins dimerization.30 The functions of circRNAs are only partially characterized. CircRNAs have been described as super-spongers, being able to bind tens of miRNA molecules and inhibit their function.31,32 However, only a few circRNAs are capable of binding multiple miRNA molecules.29 Similar to lncRNAs and immature miRNAs, circRNAs can code for micropeptides.1 Additionally, circRNAs bind proteins33 and regulate their functions and can control translation29 (Fig. 1c). These functions are seen more as exceptions than rules, and the mechanistic roles of circRNAs need to be further researched. Their role in cancer was initially revealed through deep sequencing profiling when it was observed, in 2013, that many circular transcripts are abundant and differently expressed in multiple cancer cell lines.34 Soon after, this observation was confirmed in patients’ samples.29
The three ncRNA classes have been extensively linked to different malignant processes, including resistance to various cancer therapies. Interesting is the fact that the same miRNA was shown to be an oncogene in one cancer and a tumor suppressor gene in another cancer.6 Hence, miRNAs play a context-dependent role in tumorigenesis. LncRNAs are known to regulate all hallmarks of cancer, and because of their 3D structure, single nucleotide polymorphisms and mutations can induce important functional switches that have only recently started to be characterized.35 CircRNAs are the “newest” addition and have also been linked to all cancer hallmarks, their function in cancer being mainly explained by miRNA sponging.29 Indeed, all three classes of ncRNA directly or indirectly interact—lncRNAs and circRNAs can bind miRNAs and inhibit their binding to mRNAs—so a complex network of RNA molecules exists. In order to discover crucial targets that could reverse therapy resistance in cancer, this network’s essential hubs need to be revealed.
In recent years, we have witnessed multiple high-throughput studies (e.g., genome sequencing, transcriptomics, proteomics) researching the role of mutational, transcriptional, and translational aberrations in drug resistance.36 Nevertheless, a thorough understanding for lack of response to therapy in many instances has not yet been found. We suggest that the constantly increasing number of ncRNAs—which includes other species such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), piwi-interacting RNA (piRNAs), small nuclear RNA (snRNAs), and small nucleolar RNAs (snoRNAs), not discussed here for the lack of space and because they are beyond the scope of this paper, but reviewed by others37—could be the missing elements needed to understand therapy resistance. First, ncRNA levels change quickly and are extremely heterogeneous between tumors with similar histological subtypes. This makes ncRNAs difficult to use as screening and diagnostic biomarkers but interesting biomarkers for sub-classifying a tumor type and hence useful tools for personalized medicine. These quick changes in expression (bursts) that we observe, which in many cases are from undetectable (i.e., not expressed) to highly expressed, can explain the phenomenon of acquired resistance—which sometimes takes place quickly and is hard to understand in the context of slow events having a complex mechanism of occurrence, such as mutations, translational changes, or epigenetic alterations. Second, ncRNAs are extremely versatile. The three classes of ncRNAs have multiple functions, and the phenomenon of resistance can emerge not by changes in transcription level but by changes in function. “Functional switches” are not well studied in the context of therapy resistance but are well documented in various pathological mechanisms for all three types of ncRNAs. The best studied functional switches are those for miRNAs; for example, miR-21-5p can bind TLR8 and induce a protumorigenic inflammatory response,38 and pri-miR-200a and -200b can be translated into micropeptides that inhibit epithelial-mesenchymal transition.6 This shows that in some instances, no change in expression is necessary for inducing phenotypical changes, but only a functional switch. Functional switches are most probably dependent of the subcellular localization of ncRNAs, and we believe that a better understanding of such mechanisms will be achieved with the development of spatial transcriptomics for ncRNAs. Finally, as we already mentioned, there is a complex interplay between the different classes of ncRNAs as each type of ncRNA can bind any other type, creating intricate networks,39,40 and a change in one ncRNA can induce a domino effect that can modify a vast number of molecules.
Hence, we consider ncRNAs to be potential markers that can predict a personalized response to therapy or even adjuvants that can increase response to conventional therapy. In the next section, we will present some prominent examples of ncRNAs that play important roles in therapy resistance.
Mechanisms of therapy resistance mediated by ncRNAs
Treatment resistance41 can be classified as intrinsic or acquired according to the timepoint when the resistance develops. Intrinsic resistance is the innate resistance that exists before the initiation of treatment or develops within a short duration after treatment initiation. Intrinsic resistance represents a lack of response to the initial treatment. Acquired resistance occurs after a certain duration of the treatment.42 In this scenario, the cancer initially responded to treatment but later progressed.
Intrinsic resistance usually is caused by the following mechanisms: (1) innate genetic aberrations leading to the poor response to various cancer therapies, e.g., NSCLC with EGFR (epidermal growth factor receptor) T790M de novo mutation has no response to first- and second-generation EGFR tyrosine kinase inhibitors (TKIs)43 and breast cancer with absence of estrogen receptors or progesterone receptor does not benefit from endocrine therapy;44 (2) heterogeneity within tumors tissues in which pre-existing resistant subpopulations will survive anti-cancer treatment, e.g., cancer stem cells with the capacity of self-renewal and differentiation will survive and contribute to tumor repopulation and growth;45 (3) protection induced by the activation of defending intrinsic pathways against xenobiotics, e.g., activation of ATP-binding cassette efflux transporters or the glutathione/glutathione S-transferase system to cause the efflux of chemotherapeutic drugs.46
There are also multiple mechanisms of developing acquired resistance: (1) driver oncogene modification, e.g., development of EGFR T790M mutation, but not de novo alteration, is observed within 1 year in about 50% of NSCLC patients treated with the first and second generations of TKIs, resulting in tumor progression;43 (2) activation of independent pro-survival parallel signaling, e.g., cell proliferation, apoptosis, or autophagy and cell metabolism signaling;47 (3) adaption of the tumor microenvironment after the start of treatment. Of note, these mechanisms of developing the intrinsic and acquired resistance usually co-exist and contribute to tumor progression; thus, it is more practical to understand the exact underlying mechanisms of resistance development than to seek insight into intrinsic and acquired resistance separately. NcRNAs directly or indirectly modulate the treatment sensitivity by finely orchestrating these underlying mechanisms. Overexpression of some lncRNAs can function as tumor driver oncogenes to promote the intrinsic chemotherapy resistance, while others are overexpressed after the induction of treatment and then modulate survival signaling to promote tumor repopulation, leading to acquired resistance. This section will give a short outline of the roles of the most studied ncRNAs in intrinsic or acquired therapeutic resistance and their potential mechanisms.
Resistance to chemotherapy
Many factors can induce chemotherapy resistance, but probably the most important is tumor heterogeneity.48 For intrinsic resistance, intertumoral heterogeneity plays a crucial role, and the genetic variability (germline variations) between patients harboring neoplasia of the same histotype explains why only some tumors will respond to a given chemotherapy agent. For acquired resistance, spatial and temporal intratumoral heterogeneity is the key element,48 and it is accepted that chemotherapy induces the selection of tumor cell populations that are resistant. One of the crucial mechanisms behind intratumoral heterogeneity is chromosomal instability (CIN), the continuous duplications and deletions of chromosomal regions during cancer cell division (Box 1).
For almost a decade, it was known that CCAT2, an lncRNA located in the frequently amplified 8q24 region, is overexpressed in colorectal cancer (CRC) and is associated with CIN.49 Recently, the molecular mechanism related to its role in CIN was revealed. CCAT2 binds BOP1 and AURKB, two proteins known to be associated with CIN, and increases the number of chromosomal aberrations. As expected, this induces abnormal mitosis in vitro and in vivo. Not surprisingly, high CCAT2 levels in CRC cell lines are associated with resistance to the two main chemotherapeutics used in gastrointestinal cancers, 5-flurouracil (5-FU) and oxaliplatin.50 By studying the role of mesenchymal stem cells (MSCs) in gastric cancer, He et al. proved that their role in chemoresistance is mediated by the lncRNA MACC1-AS1. The researchers discovered that MSCs induce stemness and chemotherapy resistance by secreting transforming growth factor β1, which in gastric cancer cell lines induces overexpression of SMAD2 and SMAD3, which in turn activate MACC1-AS1 expression. MACC1-AS1 binds and inhibits miR-145-5p, derepressing to key elements (CPT1 and ACS) of the fatty acid oxidation pathway. In vivo experiments reveled that inhibition of the fatty acid oxidation pathway restored gastric cancer sensitivity to the FOLFOX regimen, which includes 5-FU, oxaliplatin, and folinic acid.51
By comparing cisplatin-resistant with cisplatin-sensitive bladder cancer cell lines, Drayton et al. detected a signature of dysregulated miRNAs that is associated with resistance development. To better characterize the resistance mechanism, the authors analyzed whether the resistance is mediated by cellular metabolic changes prior to DNA adduct formation or via DNA damage repair mechanism after adduct formation. Unexpectedly, they observed that cisplatin resistance in bladder cancer is induced by an altered cisplatin metabolism in which production of glutathione and SLC7A11 are increased. It is important to mention that intracellular glutathione binds cisplatin and detoxifies the intracellular environment. One of the miRNAs found downregulated in the initial screening, miR-27a, directly binds SLC7A11 and decreases glutathione production. Hence, low levels of miR-27a are responsible for cisplatin resistance in bladder cancer. Finally, in clinical samples, the authors confirmed that high levels of SLC7A11 and low levels of miR-27a are associated with poor prognosis.52
In an attempt to understand the mechanism of cisplatin resistance in gastric cancer, it was observed that patients who acquired resistance had a significantly higher level of circAKT3, and high circAKT3 was associated with shorter overall survival. Indirectly, it was observed that high levels of circAKT3 increase the level of genomic instability by interfering with the DNA damage repair protein BRCA1. Additionally, circAKT3 inhibits the function of miR-198, which depresses the oncoprotein PIK3R1, which in turn activates the well studied PI3K/AKT oncogenic pathway.53
Resistance to targeted therapy
Targeted therapy development was possible due to the evolution from an empirical-based drug discovery approach to a rational approach in which an aberrant dominant mutation, gene amplification, or oncogenic translocation that drives tumor growth is targeted.54 One characteristic of targeted therapy, especially for solid tumors, is that only a minority of tumors rely on the hyperactivation of the targeted genes to evolve.54 In patients with intrinsic resistance, targeted therapy will not be started because molecular analysis shows that the drivers are missing. In patients who are candidates for targeted therapy, response is usually not permanent but temporary. After the initial response phase, acquired resistance develops.
A commonly used targeted therapeutic agent, sunitinib, is a TKI approved for the treatment of gastrointestinal stromal tumors (GISTs), pancreatic neuroendocrine tumors, and renal cell carcinomas (RCCs). Unfortunately, up to 20% of patients with RCC show an intrinsic resistance to sunitinib, and most of the other patients develop resistance during the course of therapy.55 Qu et al. used in vitro and in vivo screening algorithms to discover new pathways associated with sunitinib resistance.56 They observed that a previously uncharacterized lncRNA, lncARSR (lncRNA activated in RCC with sunitinib resistance), is upregulated after resistance development. Using multiple clinical samples, they observed that the level of circulating lncARSR in plasma was higher in patients with progressive disease and that high levels were associated with shorter overall survival. Mechanistically, it was observed that the RNA binding protein hnRNPA2B1 packs lncARSR into exosomes, and these are transferred between cells, disseminating sunitinib resistance. Moreover, by injecting exosomes from sunitinib-resistant cells into naïve tumors of mice, they induced sunitinib resistance in vivo. They showed that at the intracellular level, lncARSR binds miR-34a and miR-449, indirectly upregulating AXL and c-MET. Finally, in a proof-of-concept experiment, the authors restored sunitinib resistance in vivo by targeting lncARSR using a complementary locked nucleic acid inhibitor.56
After establishing a 3D model of resistance to the EGFR inhibitor cetuximab, Lu et al. discovered that the most notable transcriptional event acquired by the newly developed model was an upregulation of MIR100HG primary transcript and the two mature hosted miRNAs, miR-100 and miR-125b. Phenotypically, the two miRNAs additively play an oncogenic role and mediate cetuximab resistance in vitro and in vivo. Mechanistically, miR-100 and miR-125b inhibit five negative regulators of the Wnt signaling pathway: DKK1, DKK3, ZNRF3, RNF43, and APC2, hence stimulating this pro-oncogenic circuit. The upstream expression of the lncRNA MIR100HG is negatively regulated by the GATA6 transcription factor, which is downregulated in cetuximab-resistant and advanced stage CRC. Moreover, miR-125b binds the 3’UTR of GATA6, inducing its post-transcriptional inhibition and creating a double negative feedback circuit. Clinical data showed an important increase in MIR100HG and its embedded miRNAs and a decrease in GATA6 at the time of disease progression during cetuximab treatment.57
Sorafenib is a multi-kinase inhibitor approved for the treatment of advanced RCC, hepatocellular carcinoma (HCC), and thyroid cancers. A significant number of patients with HCC respond poorly to sorafenib, while responders frequently develop resistance during the first 6 months of therapy.58 Starting from the observation that high miR-541 levels are associated with longer overall survival in HCC, Xu et al. study the anti-oncogenic function of this miRNA. miR-541 directly targets Ras-related protein RAB1B and autophagy-related gene 2 A (ATG2A), strongly inhibiting autophagy both in vitro and in vivo. More remarkable is the fact that high levels of miR-541, in an additive manner, potentiate the anti-tumorigenic effect of sorafenib. This phenomenon is most probably mediated via inhibiting RAB1B and ATG2A. Clinical data strongly support these findings; patients with a high level of miR-541 who were treated with sorafenib had significantly longer survival compared to patients with high miR-541 and without sorafenib therapy.59 Another study by Xu et al. showed that circRNAs also can influence resistance to sorafenib. CircRNA-SORE (a circRNA upregulated in sorafenib-resistant HCC cells) not only is upregulated in multiple sorafenib-resistant cell lines but is a key element in maintaining that resistance. At the molecular level, circRNA-SORE directly binds in the cytoplasm the oncogenic protein YBX1 and prolongs its half-life by blocking its transfer into the nucleus where it is degraded by PRP19. Similar to lncARSR, circRNA-SORE is transferred from resistant cells to naïve cells via exosomes and induces a widespread resistance to sorafenib. By treating mice bearing subcutaneous sorafenib-resistant patient-derived xenograft tumors with small interfering RNA (siRNA) against circRNA-SORE, the authors showed that inhibition of the circRNA can restore sorafenib resistance.60
Resistance to radiotherapy
It is accepted that radioresistance is controlled by intrinsic factors arising from tumor cells, mainly the genomic instability characteristic for many neoplasia,61 or by extrinsic factors represented by multiple components of the tumor microenvironment (i.e., the immune component, vascular component, and pro-fibrotic stromal component).62
Starting from the observation that linc00312 is downregulated in nasopharyngeal carcinoma compared to chronic rhinitis, Guo et al. studied its role in cancer. They discovered that this lncRNA is much higher in radiotherapy-treated patients with complete response compared to those with partial response and progressive disease/radioresistance. In vitro experiments confirmed the tumor suppressor function of nuclear linc00312, which inhibits proliferation, activates apoptosis, and renders radiosensitivity to cancer cells. At a molecular level, linc00312 directly binds the catalytic subunit of DNA-dependent protein kinase, inhibiting its interaction with the Ku80 subunit after DNA double-strand breaks. Hence, it seems that linc00312 potentiates radiotherapy by blocking the DNA repair machinery.63
By comparing patients with breast cancer whose disease relapsed after radiotherapy versus those whose disease did not relapse, it was observed that a panel of miRNAs is dysregulated. In particular, miR-139-5p was downregulated in patients with unfavorable outcomes, and its overexpression was associated with high sensitivity to radiotherapy in vitro. Mechanistically, it was observed that this miRNA targets multiple genes with important roles in DNA repair and reactive oxygen species (ROS) defense, including MAT2A, POLQ, TOP1, and TOP2A. By overexpressing miR-139-5p in radiotherapy-resistant cells, the DNA repair mechanism was blocked and apoptosis induced. Using a massive patient cohort, it was confirmed that high levels of miR-139-5p and low levels of POLQ, TOP1, and RAD54L are associated with better survival, but only in radiotherapy-treated patients. Finally, by using miR-139-5p mimetics in a proof-of-concept experiment in vivo, it was proven that miR-139-5p is a potent radiotherapy sensitizer.64
Yuan et al. discovered that high levels of miR-410 induce radiotherapy resistance in NSCLC by accelerating DNA damage repair. At the molecular level, miR-410 directly binds and inhibits the translation of the tumor suppressor PTEN, which in turn activates the PI3K/mTOR signaling pathway. Moreover, miR-410 also activates epithelial-mesenchymal transition (EMT) via the PI3K/mTOR signaling pathway. Clinical observations confirmed these findings: miR-410 is overexpressed in EMT and mesenchymal tumors and is associated with low levels of PTEN.65
The paradigm regarding the meaning of non-coding is shifting. Recently it was shown that in glioblastoma multiforme, the levels of the already mentioned circAKT3 drop. But much more surprising, this circRNA encodes protein AKT3-174aa, which is 174 amino acids long and plays important anti-tumorigenic roles. AKT3-174aa interacts with the RTK/PI3K/AKT pathway, inhibiting the phosphorylation of AKT at Thr308. From a therapeutic standpoint, AKT3-174aa overexpression restored glioblastoma cells’ sensitivity to radiotherapy. Therefore, we can envision in the near future the delivery of ectopic proteins/peptides encoded by ncRNAs as new adjuvants to restore sensitivity to radiotherapy.66
Resistance to immune checkpoint inhibitors
Immune checkpoint inhibitors (ICIs), monoclonal antibodies directed against immune checkpoint molecules such as PD-1, PD-L1, and CTLA-4, are the newest addition to cancer therapy. These drugs are true game changers of cancer therapy, inducing durable disease control and prolonged response. Unfortunately, not all treated patients experience effective responses.67 Mechanisms of resistance to immune checkpoint therapy can be divided into (1) deficient anti-tumor T cell production, (2) poor anti-tumor T cell effector function, and (3) impaired development of T cell memory.68 Additionally, resistance to ICIs was linked to other immune cells such as natural killer (NK) cells and myeloid-derived suppressor cells (MDSCs).
Starting from the observation that the lncRNA LINK-A is overexpressed in patients whose disease does not respond to pembrolizumab and has a negative correlation with CD8+ T lymphocyte and antigen-presenting cell expression, Hu et al. described the function of this lncRNA in the intrinsic resistance to ICI. The authors used an existing breast cancer mouse model in which they overexpressed LINK-A and discovered that it induces an aggressive triple-negative breast cancer phenotype that metastasizes to the lungs. Mechanistically, LINK-A facilitates the interaction between phosphatidylinositol-(3,4,5)-trisphosphate (PtdIns(3,4,5)P3) and G-protein–coupled receptor, decreasing the phosphorylation of TRIM71. An outcome of this interaction leads to increased degradation of TP53, Rb, and the antigen peptide-loading complex. Furthermore, this molecular cascade decreases the number of CD8+ T cells and granzyme B NK cells in the peritumoral milieu.69
An additional element associated with ICI resistance is MDSCs, high levels of which may be associated with resistance to ICIs.70 Huber et al. discovered that multiple miRNAs, miR-146a, miR-155, miR-125b, miR-100, let-7e, miR-125a, miR-146b, and miR-99b, are released by melanoma cells via extracellular vesicles (EVs). Consequently, EVs containing this set of miRNAs are internalized into myeloid cells, which in turn acquire an MDSC phenotype. Clinical data revealed that in patients with stage IV melanoma treated with the ICIs nivolumab or ipilimumab, high levels of this set of circulating miRNAs are associated with shorter overall survival.71 Hence, we can envision combining ncRNA therapy with ICIs to overcome resistance (Box 2).
Huang et al. adopted a classic method to study therapy resistance in HCC; they started by analyzing genes located in the 7q21-7q31 amplicon associated with an unfavorable outcome. They observed that circMET is located in this region, is overexpressed in HCC, and is associated with unfavorable outcomes. At a phenotypical level, they noticed that circMET overexpression induces EMT and potentiates the immunosuppressive tumor microenvironment. Immunologically, circMET decreases the density of CD8+ lymphocytes in tumor tissue. At the molecular level, circMET sponges miR-30-5p and indirectly upregulates the transcription factor Snail. Snail activates the expression of DPP4, which in turn inhibits the chemotactic molecule CXCL10, hence blocking CD8+ immune cell trafficking. Finally, in vivo studies showed that if this axis is activated, anti-PD-1 therapy resistance emerges.72
An analysis of the role of another circRNA, circular ubiquitin-like with PHD and ring finger domain 1 RNA (circUHRF1), in anti-PD-1 resistance in HCC showed that NK cells also play an important role. Like circMET, circUHRF1 is overexpressed in HCC, and high levels are associated with advanced T category, decreased circulating NK cells, microvascular invasion, and short overall and relapse-free survival after surgery. Interestingly, circUHRF1 is secreted into exosomes by HCC cells, and its plasma levels are much higher before surgery and during relapse compared to after surgery or in healthy controls. At the immunological level, exosomal circUHRF1 derived from HCC cells inhibits NK cell function. In NK cells, circUHRF1 binds and inhibits the biological function of miR-449c-5p and indirectly upregulates the expression of the immune checkpoint–T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3). Further clinical analysis revealed that high circUHRF1 expression is associated with progressive disease in HCC patients treated with anti-PD-1 and negatively correlates with NK cells in tumor tissue. In vivo studies confirmed the results: mice treated with anti-PD-1 treatment plus circUHRF1 shRNA have significantly longer overall survival compared to mice treated only with anti-PD-1.73 Whether circUHRF1-mediated resistance to anti-PD-1 therapy is intrinsic or acquired needs to be further analyzed. An overview of the role of ncRNAs in therapy resistance can be found in Table 1 and Fig. 2.
Mechanisms of therapy resistance mediated by ncRNAs. Examples of the common mechanisms of cancer cell resistance to tyrosine kinase inhibitors, chemotherapy, radiation, and immune checkpoint inhibitors mediated by miRNAs, lncRNAs, or circRNAs. The common mechanisms include (1) modulation of defending intrinsic pathways against the xenobiotics, e.g., miR-27a directly binds to SLC7A11 and decreases the glutathione (GSH), which binds cisplatin and detoxifies the intracellular environment, thus a decrease of miR-27a is responsible for cisplatin resistance; (2) promoting survival signaling pathways, e.g., lncARSR, which is packed by hnRNPA2B1 and then binds to miR-34a and miR-449, indirectly upregulates AXL and c-MET to contribute to sunitinib resistance; MIR100HG and its embedded miRNAs, miR-100 and miR-125b, mediate cetuximab resistance by activating Wnt signaling; circRNA-SORE directly binds to oncogenic protein YBX1 and prolongs its half-life by blocking its transfer into the nucleus, where it is degraded by PRP19 to trigger sorafenib resistance; (3) accelerating DNA damage repair, e.g., miR-410 inhibits the translation of PTEN, leading to the activation of the PI3K/mTOR signaling and accelerating DNA damage repair to induce radiotherapy resistance; circAKT3 inhibits miR-198, which in turn activates the PI3K/AKT signaling and triggers cisplatin resistance; (4) inducing genomic instability, e.g., lncRNA CCAT2 binds with BOP1 and AURKB to induce chromosomal instability (CIN) and resistance to 5-flurouracil (5-FU) and oxaliplatin; (5) inhibition of cell apoptosis or autophagy, e.g., miR-541 targets Ras-related protein RAB1B and autophagy-related gene 2 A (ATG2A), inhibiting autophagy, and further accelerates sorafenib resistance; (6) regulating cell metabolism, e.g., MACC1-AS1 binds and inhibits miR-145-5p, derepressing to key elements (CPT1 and ACS) of the fatty acid oxidation pathway, leading to resistance to the FOLFOX chemotherapy regimen; and (7) tuning the infiltrated immune cells, including T cells, myeloid-derived suppressor cells (MDSCs), and natural killer cells in the tumor immune microenvironment, e.g., circMET sponges miR-30-5p and indirectly inhibits the chemotactic molecule CXCL10, hence blocking CD8+ immune cell trafficking; LINK-A facilitates the degradation of TP53 and Rb, thus decreasing the number of CD8+ T cells and granzyme B NK cells; circUHRF1 binds and inhibits miR-449c-5p, upregulating TIM-3, to inhibit NK cell function; and miR-146a, miR-155, miR-125b, miR-100, let-7e, miR-125a, miR-146b, and miR-99b are released by melanoma cells via extracellular vesicles and internalized into myeloid cells to drive MDSC differentiation.
Non-coding RNAs as biomarkers for therapy resistance
Biomarkers are regarded as signs of a biological process, indicating a certain condition or disease, and are usually assessed, invasively or non-invasively, from body fluids or tissues.74 A feasible cancer biomarker is the one that is expressed by a specific type of cancer cell, differentially expressed compared to normal tissue, or dynamically altered during cancer progression or the course of treatment.75 Such biomarkers have assumed a growing role in distinguishing malignant from benign disease, predicting patient prognosis, monitoring cancer recurrence, and determining response to anti-cancer therapy. Prominent biomarker candidates are identified as proteins (i.e., cytokines and receptors) and nucleic acids (i.e., DNA, RNA), including ncRNAs.76 Vast evidence reveals that some ncRNAs are the preferential biomarker in the diagnosis of certain cancers, especially when comprehensively combined with other biomarkers.77,78 Tissue-specific, cell-specific, and developmental stage–specific expression patterns give ncRNAs great value as clinical biomarkers in certain cells, tissues, and conditions. By annotating the gene expression of 16 tissues through GENCODE consortium, the expression patterns of 14,880 lncRNAs were revealed. Compared to protein-coding genes, 65% of which were detected in all human tissues, only 11% of lncRNAs were detected in these tissues, which suggested that lncRNAs show more tissue-specific expression patterns.79 The expression of lncRNA in T cell lineages is a good example of its cell- and developmental stage–specific expression patterns. Hu et al. conducted a pair-wise comparison of protein-coding genes and lncRNAs between different stages of T cell development. Their results indicated that mRNAs are similarly expressed between different T cell subsets, while remarkably different lncRNAs were expressed between various T cell subsets. Quantitative analysis showed that 48–57% of lncRNAs, in contrast to 6–8% of coding genes, were specifically expressed in various T cell subsets.80 This was further proved by other studies. By profiling lncRNA expression of CD8+ T cell subsets in both humans and mice, researchers found that lncRNA-Snhg1, which exhibits the naivehi-effectorlo-memoryhi expression pattern, plays an essential role for memory CD8+ T cell establishment. Thus lncRNA-Snhg1 could be a unique biomarker to identify this subset of T cells.81 Certain ncRNAs are also candidate biomarkers for predicting therapy resistance.
Considerable attention has been paid to the use of non-invasive methods such as liquid biopsies to analyze biomarkers from body fluids (e.g., blood, saliva, urine). The reliability and reproducibility of these assays to detect and characterize tumors have tremendous value with far-reaching clinical implications. The use of biomarkers in body fluids to predict cancer therapy response has made significant progress, allowing for the selection of appropriate treatment options.82 Biomarkers that are easily accessible from body fluids are circulating tumor cells, circulating proteins, DNA, and RNA, including ncRNAs. Circulating RNAs are largely secreted by cells and therefore give hints regarding diseases and biological processes, including response to therapy. Harnessing the role of certain ncRNAs in intrinsic and acquired treatment resistance has led to their study as biomarkers that can predict therapeutic outcomes in a given patient before, during, or after treatment. This association is partially dependent on the property of ncRNAs to function in cell-to-cell communication, mediating drug resistance.83
NcRNAs can travel in body fluids in three different forms: bound to proteins, bound to lipoproteins, or inside small EVs. The mechanisms are especially well described for miRNAs. NcRNAs form RNA-protein complexes, can be released by cells, and are probably the predominant mechanism of cell-to-cell communication. Argonaute complexes, the pivotal component of the miRNA-induced silencing complex formed inside cells, contribute to the stability of plasma miRNAs by binding them.84 Lipoproteins such as high- and low-density lipoproteins (HDL and LDL) are inherently soluble and have the tendency to embed water-insoluble material inside their core, which enables them to transport nucleic acids between cells and also protects miRNAs from degradation by RNases.85,86 These miRNAs are then transferred to recipient cells and can regulate downstream gene expression. Another interesting form of cell-to-cell communication is mediated by EVs. Exosomes, the smallest subclass of EVs, have been extensively investigated recently in cancer pathogenesis.87 They are produced via exocytosis of multivesicular bodies that enclose various types of molecules, including ncRNAs, and are secreted into the interstitial spaces circulating in body fluids. By endocytosis of ncRNAs enclosed into exosomes of neighboring or remote recipient cells, cell signals can be transferred between cells, including the drug-resistant phenotype.88
The role of ncRNAs as critical regulators of carcinogenesis and therapeutic resistance is supported by in vivo and in vitro data, and the focus of this section is to discuss ncRNAs as biomarkers to predict response in cancer therapy.
Chemotherapy resistance
Multidisciplinary cancer treatment is being effectively used worldwide. Though chemotherapy is one of the traditional standard approaches for cancer management, only a fraction of patients will experience objective clinical response to various chemotherapy regimens. Therefore, characterizing novel biomarkers to discriminate patients who are intrinsically resistant to the planned chemotherapy will avoid unnecessary adverse side effects. 5-FU and oxaliplatin are two fundamental chemotherapy agents that are components of the most common chemotherapy regimens for CRC and other gastrointestinal cancers, e.g., FOLFOX, FOLFIRI, and XELOX. By screening the differentially expressed miRNAs from 20 matched CRC serum samples with or without objective response to oxaliplatin-based chemotherapy, Zhang et al. identified five miRNAs—miR-20a, miR-130, miR-145, miR-216, and miR-372—that were significantly downregulated in responders compared to non-responders. The area under the receiver operating characteristic curve (AUC) values of this group of miRNAs in the training and validation set comprising of 40 and 173 samples were 0.841 (95% CI: 0.707–0.975) and 0.918 (95% CI: 0.871–0.963), respectively. This miRNA signature also demonstrated better accuracy in predicting chemotherapy resistance than traditional tumor biomarkers such as CEA (AUC = 0.689, 95% CI: 0.618–0.0.760), and CA19-9 (AUC = 0.746, 95% CI: 0.682–0.851).89 However, whether the serum samples were obtained before or after the initiation of the treatment is unclear.
Similarly, patients with metastatic CRC who were resistant to first-line 5-FU/oxaliplatin-based chemotherapy showed higher expression of miR-130b, miR-106a, and miR-484 compared to responders. The data were further validated in another cohort of 150 patients.90 Of note, the plasma samples were obtained prior to treatment, suggesting that these plasma miRNAs may serve as non-invasive markers to predict intrinsic resistance to 5-FU and oxaliplatin–based chemotherapy in metastatic CRC patients. In another study, of 742 miRNAs profiled in metastatic CRC patients who did and did not respond to XELOX/FOLFOX, high expression of miR-625-3p was correlated with poor response; this finding was validated in a cohort of 94 patients (OR = 6.25, 95% CI:1.8–21.0). However, miR-625-3p was not associated with prognosis, suggesting that miR-625-3p might solely be a response-predicting biomarker. miR-625-3p was also overexpressed in an oxaliplatin resistance–induced HCT116 cell line compared to parental cells.91 MAP2K6-p38 signaling might be involved in the induction of this resistance.92
MiR-20a, miR-145, and miR-106a are also widely acknowledged as key miRNAs in chemotherapy resistance.93,94 miR-20a-5p regulates chemosensitivity to gemcitabine by targeting ribonucleotide reductase subunit M2 in pancreatic cancer and predicts the response to gemcitabine-based chemotherapy with satisfying predictive value (AUC = 0.89).95 Upregulation of miR-20a and downregulation of miR-451 after the second cycle of neoadjuvant chemotherapy, which is widely applied to treat locally advanced breast cancer, predicted resistance to treatment in HR+/HER2- breast cancer (AUC = 0.80 and 0.788, respectively).96 Though miR-20a was associated with chemoresistance and radioresistance in in vitro and in vivo studies, these findings were not validated in independent patient cohorts.97,98,99 In a small cohort of triple-negative breast cancer patients (n = 32) who received neoadjuvant cisplatin/doxorubicin-based chemotherapy, miR-145-5p was downregulated in patients who achieved pathological complete response. The AUC of miR-145-5p as the predictor for response in this cohort was 0.7899 (95% CI: 0.6382–0.9416). It is plausible that miR-145 inhibited cell proliferation by targeting TGFβR2.100 In another study including 57 luminal breast cancer patients who received neoadjuvant chemotherapy, the level of miR-145 was significantly lower in responders compared to non-responders.101 Lim et al. performed miRNA sequencing in 1362 childhood acute myeloid leukemia samples, which comprised 1303 primary, 22 refractory, and 37 relapse samples. By applying differential expression analysis, they found that miR-106a-3p and miR-106a-5p could be biomarkers of treatment resistance, as these two miRNAs were consistently overexpressed in treatment-resistant samples—that is, refractory or relapse samples, and in primary samples from patients with induction failure. Further integrative miRNA:mRNA analysis found that miR-106a targeted the genes associated with oxidative phosphorylation, which is suppressed in treatment-resistant conditions.102 In addition, miR-9-5p, miR-9-3p, miR-433-3p, miR-21, and miR-200c may possess potentially predictive roles in chemotherapy resistance in GC and esophageal cancer (EC).103,104,105
LncRNAs also participate in the development of chemoresistance and may serve as potential biomarkers in CRC. In a cohort comprising 140 CRC patients, the lncRNA XIST was upregulated in patients who showed no response to 5-FU compared to those who showed response. These findings were validated in serum samples from 120 CRC patients from the same cohort with an AUC, diagnostic sensitivity, and specificity of 0.756, 71.7%, and 68.3%, respectively. Mechanistically, in vivo studies revealed out that XIST restrained 5-FU–induced cytotoxicity by promoting thymidylate synthase, a pivotal target of 5-FU.106 Similarly, both tissue and serum MEG3 were downregulated in oxaliplatin-resistant CRC patients. MEG3 showed potential to screen out non-responders, with an AUC of 0.784, the diagnostic sensitivity of 72.86%, and specificity of 61.43%.107 In these studies, the expression levels of lncRNAs were investigated in tissues and corresponding serum samples, demonstrating the consistency of their prognostic ability and their potential as candidate biomarkers. In a comprehensive profiling study with training and testing datasets including 1102 patients, a three-lncRNA signature (AK291479, U79293, and BC032585) was identified to predict pathological complete response after neoadjuvant chemotherapy in breast cancer.108 Liu et al. assigned different weights to the expression levels of eight lncRNAs expressed by 258 high-grade serous ovarian cancer patients from The Cancer Genome Atlas (TCGA) and successfully generated a risk-score formula for predicting chemotherapeutic sensitivity.109
Targeted therapy resistance
Angiogenesis inhibitors
MiR-126, specifically expressed in endothelial cells, plays a pivotal role in the regulation of blood vessel integrity, which might affect anti-angiogenic treatment.110 Hansen et al. reported that plasma miR-126 was dynamically increased during the treatment of patients whose metastatic CRC was resistant to first-line XELOX chemotherapy combined with bevacizumab, suggesting that miR-126 may serve as a predictive biomarker for acquired resistance to chemotherapy or bevacizumab during treatment.111 Whether chemotherapy or bevacizumab or both are regulated by miR-126 remains unknown, as anti-angiogenic therapy usually is prescribed with other combined modality therapies but not by itself. miR-126 was also reported to be involved in multi-drug resistance through a variety of mechanisms, e.g., contributing to sorafenib resistance.112 miR-126 is also well known for endowing leukemia stem cells with chemotherapy resistance ability. It was significantly upregulated in relapse blasts compared to paired diagnostic samples and also after induction or salvage chemotherapy in acute myeloid leukemia patients.113 In contrast, there was an inverse correlation between the level of miR-126 and acquired resistance to dabrafenib in melanoma and tamoxifen treatment in estrogen receptor-positive breast cancer, suggesting multiple roles for the same miRNA in different therapies.114,115 Rinnerthaler et al. divided two cohorts of breast cancer patients treated with chemotherapy with or without bevacizumab into responder and non-responder groups according to the length of progression-free survival and then selected the differentially expressed miRNAs between the two groups. By identifying the mutually differentially expressed miRNAs and the miRNAs with prognostic power from these two cohorts they selected 12 miRNAs that provide survival information. Finally, in a validation cohort of 230 patients from a randomized trial, they confirmed that low expression of miR-20a-5p was the only predictor of benefit from bevacizumab-containing therapy.116 Interestingly, decreased expression of the same miRNA, miR-20a, in CRC positively correlated with treatment response with oxaliplatin-based chemotherapy, indicating miRNA specificity for treatment and disease state,89 which could be attributed to molecular mechanisms that govern the disease and site of action. However, predictive measures such as the AUC, sensitivity, and specificity of using these miRNAs need to be further investigated.
Tyrosine kinase inhibitors
The unique histological and molecular features of lung cancer, especially NSCLC, have offered considerable promise for precise personalized medicine in multidisciplinary cancer management. This has been made possible because of tremendous efforts that unraveled the underlying molecular mechanisms, particularly the discovery of mutations and/or alteration of genes such as EGFR, ALK, and ROS1. Despite EGFR TKIs’ selectively targeting EGFR-mutant NSCLC with significant treatment response, 20–30% of patients either do not respond or respond for less than 3 months; these are considered to have intrinsic resistance to treatment.43 By profiling the different miRNAs in gefitinib-sensitive and -resistant samples with EGFR mutation, miR-25, miR-122, miR-195, miR-21, and miR-125b were identified to predict gefitinib sensitivity in EGFR-mutated NSCLC.117 The AUC (0.869) of the combination of these plasma miRNAs had shown a discriminatory power of detecting EGFR mutation. This could be an indication for using plasma EGFR analyses of cell-free DNA when it is infeasible to get tissue samples to detect EGFR mutation status. However, no validation of the predictive value of this panel miRNAs in predicting the intrinsic resistance for EGFR-TKI was performed. Furthermore, except for the primary and secondary T790M mutation, mechanisms contributing to the resistance of EGFR-TKI have not been fully explored. Besides its role in inducing oxaliplatin resistance in CRC, miR-625-3p was also reported to induce a T790M-indepedent acquired resistance by activating the TGF-β/Smad pathway and EMT in vitro.118
Secondary imatinib resistance is the major reason for therapeutic failure in GISTs and poses a huge clinical challenge. The level of serum miR-518e-5p is higher in patients with GIST and secondary imatinib resistance than those with imatinib-sensitive GIST. The AUC, sensitivity, and specificity of miR-518e-5p to predict response to imatinib were 0.9938, 99.8%, and 82.1%, respectively, which demonstrated a satisfactory ability to discriminate the resistant tumors.119 Around 65% of patients have intrinsic resistance to bortezomib and do not respond to treatment with this widely used targeted therapy for multiple myeloma.120 The integrated expression of miR-215-5p, miR-181a-5p, and miR-376c-3p, with an AUC of 0.95 (95% CI: 0.84–1.00), could discriminate between patients with refractory versus sensitive multiple myeloma treated with bortezomib.121 The miRNA signature model identified in this study could serve to enhance the rate of treatment success.
Radiotherapy resistance
EC patients who cannot undergo esophagectomy receive concurrent chemoradiotherapy as the alternative standard treatment, but only 30–50% achieve a permanent response. Radioresistance has been implicated in the upregulation of miR-193b, which increases the proportion of cells in the G0/G1 phase. Serum miR-193b was significantly lower in patients who had a complete response than in those who exhibited a partial response after radiotherapy, and it had a good predictive value for detecting EC patients who achieved a complete response (AUC = 0.710, 95% CI: 0.580–0.839).122 Besides the expression level itself, single nucleotide polymorphisms of ncRNAs or associated regulatory regions also correlated with radiosensitivity. For example, rs4938723 in the promoter region of miR-34b/c was related to chemoradiotherapy response in EC. Data from 175 patients showed that patients with the CC rs4938723 genotype had a better response to chemoradiotherapy than that of patients with TT or TC genotypes. The predictive model showed an AUC, sensitivity, and specificity of 0.777, 85.1%, and 71.3%, respectively, which was considered promising for EC patients.123
In patients with locally advanced rectal adenocarcinoma, an lncRNA signature comprising lnc-KLF7-1, lnc-MAB21L2-1, and LINC00324 was validated to predict the response to neoadjuvant chemoradiotherapy, with good performance (AUC = 0.93).124
Immunotherapy resistance
Identifying clinical biomarkers that can accurately predict the response to immunotherapy remains a significant challenge for the widespread application of ICI. Depending on the type of ICIs, immunohistochemistry expression of PD-L1/PD-1 on tumor cells and immune cells and tumor mutation burden (TMB) have emerged as promising biomarkers for predicting response to immunotherapy. Expression of PD-L1 by immunohistochemistry in tumor samples was approved by the US Food and Drug Administration (FDA) to be the criteria for the use of some ICIs, e.g., the indication for pembrolizumab in treating metastatic NSCLC. However, the requirement of biopsies and imprecise assessment of the results due to the intratumor heterogeneity limits its application. Thus, ncRNAs, especially the circulating ones that directly and indirectly target immune checkpoint molecules such as PD-1/PD-L1, TIM3, CTLA-4, B7-H3, and LAG-3, can be also implicated as biomarkers with great potential.125,126,127 There is a correlation between high TMB and response to ICIs in microsatellite instability high metastatic CRC.128 High TMB represents a high abundance of neo-epitopes that arise from the modification of proteins encoded by mutated genes, which leads to the activation of anti-cancer immune responses against those neoantigens. The survival of patients with head and neck squamous cell carcinoma has recently been prolonged with the implementation of ICIs. Therefore, Xia et al. explored whether a 25-miRNA-based classifier from the head and neck squamous cell carcinoma cohort in the TCGA database can predict TMB levels to identify patients who truly benefit from ICIs. The AUCs of this 25-miRNA-based signature model to predict TMB status were 0.822 for the training set, 0.702 for the test set, and 0.774 for the total set.129 Similarly, in uterine corpus endometrial carcinoma, the AUCs of a 26-miRNA signature for predicting TMB were 0.869 for the training set, 0.904 for validation the set, and 0.820 for the total set. This miRNA signature pattern also correlated with the expression of PD-1 and PD-L1, mismatch repair-related genes such as MLH1 and MSH6, and homologous recombination repair of double-strand DNA break genes such as BRCA1 and BRCA2.130 A similar study was reported in lung adenocarcinoma.131 By analyzing the TCGA data for colon cancer, a multi-lncRNA signature including 14 lncRNAs for predicting TMB levels was established. This combined-classifier had better efficiency to predict TMB—with AUC levels at 0.70, 0.71, and 0.71 in three validation sets—than the traditional clinical characteristics.132 Another 33-lncRNA–based signature classifier was developed in stomach adenocarcinoma to predict TMB, with outstanding performance.133
In addition to identifying TMB, miRNAs are used as indirect biomarkers of response to ICI therapy. A phase 2 study that explored the efficacy of nivolumab, a PD-1 inhibitor, in esophageal squamous cell carcinoma revealed that serum miR-1233-5p levels (AUC = 0.895) before nivolumab treatment and miR-6885-5p, miR-4698, and miR-128-2-5p levels (AUC of 0.93, 0.97, and 0.93, respectively) after treatment initiation predicted response to ICI.134 Though this was a small study, the evidence indicates the usability of ncRNAs for future prospective clinical trials. By investigating the differences of pretreatment circulating miRNAs between responders and non-responders in patients with NSCLC who received anti–PD-1 immunotherapy, Shukuya et al. developed a response-predicting miRNA signature that consists of miR-199a-3p, miR-21-5p, and miR-28-5p. This combination had better efficiency to predict anti–PD-1 immunotherapy response—with an AUC of 0.925, which is superior to the PD-L1 expression score determined by immunohistochemistry (AUC = 0.575).135
The tumor microenvironment is populated by multiple types of immune cells: T cells, macrophages, MDSCs, and NK cells that regulate the response to immune therapy. The ncRNAs that affect the function of these essential immune cells can be implicated in predicting the response to immune therapy.136 By targeting the transcription factor T cell factor 1 (TCF1), the key regulator of effector T cells, miR-24 modulates the immune response by controlling cytokine production of T cells. Besides these, some miRNAs can be exchanged via exosomes between T cells and antigen-presenting cells during antigen recognition to mediate the immune interactions and orchestrate the immune response. It is reasonable to propose that these miRNAs can be alternative candidates to predict immunotherapy response.137
A selected list of ncRNAs with potential value in monitoring resistance to chemotherapy, radiotherapy, targeted therapy, and immunotherapy is presented in Table 2.
Therapeutic strategies to target ncRNAs to overcome therapy resistance
Various RNA-based therapies have been developed, and some have been approved by the FDA. Of note, all these therapeutics target specific mRNAs to downregulate the expression of corresponding genes. Though lncRNAs have been the focus of recent investigations, none have been clinically investigated as therapeutic targets. The utility of miRNA-based therapeutics has been developed in phase 2 and 3 clinical trials. Therapeutic modalities targeting ncRNAs are usually developed with one of two strategies: the first is to inhibit the specific ncRNA molecule if it is overexpressed, and the second is to overexpress a tumor suppressor ncRNA.138 A schematic overview of the ncRNA therapeutic strategies and delivery mechanisms is depicted in Fig. 3.
Therapeutic modalities to target ncRNAs. The therapeutic strategy to target overexpressed ncRNAs is to inhibit the specific ncRNA molecules. The inhibition modalities include (1) antisense oligonucleotides (ASOs): ASOs bind to complementary RNA sequences to block and inhibit their function and induce their degradation via RNAse-H-mediated cleavage; (2) antagomirs: antagomirs bind to complementary miRNAs and induce their degradation, thus preventing their interaction with target mRNA; (3) artificial miRNA sponges: artificial RNAs contain multiple high-affinity miRNA antisense binding sites that can sequester miRNAs from their target mRNAs; (4) small molecules: these molecules can interrupt any step of RNA transcription process; (5) small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs): these artificially synthesized double-stranded RNAs bind to complementary target ncRNA when loaded to AGO2, leading to the degradation of target RNA; (6) CRISPR/Cas9-based editing approaches, delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) to precisely cut the target ncRNA; and (7) miRNA mimics: miRNA mimics are used for replacing or substituting downregulated tumor suppressor miRNAs. Commonly used delivery systems of these ncRNA therapeutic modalities include lipid nanoparticles, exosomes, antibodies, and peptides.
NcRNA inhibitors include antisense anti-oligonucleotides (ASOs), antagomirs, siRNAs, short hairpin RNAs (shRNAs), miRNA sponges (including circRNA sponges), CRISPR/Cas9-based genome editing, and small molecule inhibitors of ncRNAs. ASOs and antagomirs are widely used inhibitors targeting miRNAs for in vitro and in vivo studies. ASOs are single-stranded RNA molecules that bind to complementary RNA sequences with well-matched base pairing to block and inhibit their function and induce their degradation via RNAse-H-mediated cleavage.139 Additionally, in preclinical in vivo studies, ASOs show specific and efficient reduction in lncRNA levels. Antagomirs are anti-miRNA ASOs that, when conjugated to cholesterol, show an improved intracellular delivery ability. Antagomirs function by complementary binding to miRNAs, thus preventing their interaction with their target genes.140 Locked nucleic acid is another commonly used chemical modification in anti-miRNA ASOs. Miravirsen (SPC3649) is a modified locked nucleic acid that is complementary to miR-122 and was investigated in phase 2 clinical trials for treating chronic hepatitis C infections.141 RNA interference is another commonly used strategy to degrade and knock down ncRNAs.142 SiRNAs are artificially synthesized double-stranded RNA molecules around 20 nucleotides long that function as RNA interference by complementary binding to their targets, leading to the transient silencing of gene expression.143 ShRNAs overcome the short lifespan of synthetic siRNAs, the main drawback, and are therefore widely employed in genetic screens and used as the common RNA interference approach in gene therapy and, occasionally, in clinical settings.144 CRISPR/Cas9 is a novel genome editing method that has been used to inhibit ncRNAs in preclinical in vivo and in vitro studies with considerable success.145 Artificial miRNA sponges are constructs containing multiple high-affinity miRNA antisense binding sites that target one specific or multiple different miRNAs.146 While the efficiency of miRNA sponging has been proven in in vivo studies, their utility in the clinic is still lacking. Small molecules either directly inhibit ncRNAs or indirectly target specific genes or proteins that regulate ncRNAs expression or function, usually involved in their biogenesis and maturation. Luciferase/GFP could be a strategy to effectively screen small molecular inhibitors that bind to mature miRNAs and block their binding to the miRNA response element.147,148 This interaction will then lead to the activation of luciferase/GFP, and the affinity could be preliminarily determined by the relative intensity of luciferase activity. Because lncRNAs exert their regulatory effect by interacting with RNA-binding proteins, small molecule inhibitors interfere with lncRNA-protein interactions and block lncRNA function.
The second strategy for targeting ncRNAs is to restore the normal function of ncRNAs that are downregulated when therapy resistance occurs. Function can be restored by replacing or substituting the lost ncRNA using synthetic ncRNA-like molecules such as miRNA mimic agents, an effective alternative widely used in in vivo studies.149 Despite remarkable progress in the field of ncRNA-based therapeutics, many challenges still need to be addressed—especially the issue of side effects caused by off-target effects.138 The well known miR-34 mimic MRX34 caused significant adverse events in five patients—with one patient suffering cytokine release syndrome—which led to the suspension of a phase 1 clinical trial for cancer treatment.15 Toll-like receptor signaling activated by miRNAs might explain this side effect: Toll-like receptors are activated, leading to the activation of downstream signaling nuclear factor-kappa B and then triggering the transcription and release of pro-inflammatory cytokines including IL-6, -8, -12, and TNF-alfa.38,150 Other non–immune-related off-target effects, due to mismatched base pairing to mRNAs that are not targets of interest, also need to be addressed. Another obstacle in RNA-based therapeutics, especially when the drug is systemically administered, is unexpected on-target effects on normal tissue but not the tumor tissue. For instance, ASO AEG35156 targeting the X-linked inhibitor of apoptosis gene also induced pathological peripheral neuropathy due to the on-target effect in neural system cells instead of cancer cells.151 Thus, the specificity, delivery, and tolerability of therapeutics using ncRNAs need to be further improved.
A safe and effective tissue delivery system for RNA-based therapeutic drugs without severe side effects remains one of the major challenges that limit their translational application. As mentioned above, antagomirs are anti-miRNA ASOs that are conjugated to cholesterol with improved intracellular entry affinity to targets. Such chemical modifications in the backbone, nucleobase, ribose sugar, and/or 2ʹ-ribose substitutions could increase the stability and efficacy of therapeutic oligonucleotides. However, an additional delivery system to enhance the affinity of the oligonucleotides is still warranted.152 Lipid nanoparticles are the most commonly used delivery system, with high biocompatibility and low toxicity.153 Liposomes are spherical nanoparticle vesicles consisting of double phospholipid layers resembling the structure of cell membranes. Liposomes are widely used to encapsulate hydrophilic or lipophilic drugs that target specific tissues.154 By avoiding the nuclease degradation and renal clearance of coated drugs, liposomes increase the cellular uptake of delivered drugs. Liposomes composed of ionizable lipid, phosphatidylcholine, cholesterol, or PEG-lipid conjugates have been successfully used in clinical trials, including the miRNA mimic MRX34 and patisiran. The latter agent is designed for treating hereditary transthyretin-mediated amyloidosis.15,155
A stimulus-responsive nanoparticle delivery system that releases the target drug in a stimuli-responsive manner (triggered by enzymes, pH, glutathione, specific temperature, hypoxia, ROS, etc.) was developed recently in order to increase the specificity of delivering the drug to the target tissue.156 This system has great advantages in increasing the homogeneous distribution and accumulation of the drug within target tumor tissue and therefore decreasing off-target effects. Moreover, physicochemical modifications to further increase its biocompatibility and spatial controls have also been employed. Gold stimulus-responsive nanoparticles, which can be easily synthesized and have a flexible size, are an ideal carrier for oligonucleotides. miR-124-5p157 and miR-145158 mimetics were recently explored to be encapsulated with these nanoparticles and shown to effectively target cancer cells. The release of these miRNA mimics is initiated by the cleavage of cystamine and then triggered by the high concentration of glutathione in the cytosol. Besides miRNAs mimics, circFoxo3 has been explored to be encapsulated within gold nanoparticles and delivered to target and induce apoptosis in melanoma cells.33 Triantennary N-acetylgalactosamine (GalNAc) can highly selectively bind to asialoglycoprotein receptor 1, which is highly expressed in the liver, making GalNAc ideal as a liver-targeted delivery system. When conjugated together with the oligonucleotides or siRNAs, GalNAc facilitates the uptake of these RNA drugs into hepatocytes by endocytosis with high selectivity.159 For example, givosiran, a GalNAc-conjugated siRNA, targets and downregulates 5ʹ-aminolevulinate synthase 1 to treat acute hepatic porphyria. Its efficacy was demonstrated in a phase 3 clinical trial.160 In addition, antibodies, aptamers, or peptides can be conjugated with siRNAs or ASOs for targeted delivery. Oligonucleotides conjugated to antibodies using click chemistry facilitate the degradation of complex after entry into the target cells, followed by releasing the ASO.161
Exosomes—EVs ranging from ~40 to 160 nm in diameter—have an endosomal origin and contain proteins, DNA, and RNA molecules. Cancer cells secrete exosomes that are delivered to distant cells to transmit intercellular messages. This intercellular communication mediated by exosomes containing ncRNAs has been proven to regulate drug resistance in various cancers.88 Because exosomes have better biocompatibility and biodistribution than synthetic delivery approaches, manipulating exosomes might hold promise as a delivery strategy in the clinic. Exosomes are natural biological nanoparticles offering unparalleled biocompatibility, and this property was harnessed in patients with severe therapy-refractory graft-versus-host disease, wherein exosomes from MSCs were safely delivered without causing severe immune reactions.162 Thus, each of the delivery strategies mentioned above could be a potential candidate for distributing ncRNA-based drugs to overcome therapy resistance. However, it should be noted that the unique design of a specific delivery system should be comprehensively based on the target ncRNA, tumor type, and other clinicopathological factors, as these features are essential to achieve the desired efficiency without causing severe off-target effects or toxicity.
Future perspectives
Several ncRNAs are being studied in clinical trials as potential biomarkers for response to cancer therapy. A curated list of such ncRNAs is provided in Table 3.
Most cancer drugs are indicated for a specific histological tissue type. Novel therapies targeting molecular aberrations in multiple cancers have been developed only recently. This concept is termed tumor-agnostic cancer treatment and represents the future of personalized cancer therapy.163 For example, the tumor-agnostic drug pembrolizumab is a monoclonal antibody against PD-1 that was initially developed for melanoma patients but has been recently approved for microsatellite instability–high and mismatch repair–deficient tumors, regardless of the cancer type.164 The TKI entrectinib was approved by the FDA 2019 to treat ROS1-positive NSCLC but also any solid cancer that has a neurotrophic tyrosine receptor kinase gene fusion.165 Of note, several ncRNAs discussed in this review are dysregulated in multiple tumor types and, therefore, may be ideal next-generation targets as tumor-agnostic ncRNA- and RNA-based therapeutics. Because of their heterogeneous expression, ncRNAs can be the ideal markers/targets for a personalized therapeutic approach that overrides our histological and mutational understanding of cancers and brings it into a non-coding transcriptional era. We envision that multiple tumor types, especially after developing a therapy-resistant phenotype, will show a dysregulated ncRNA expression pattern that will indicate to the clinician the necessity to change the treatment regimen. The ncRNAs are actively secreted in bodily fluids, and therefore, this approach can also be used as a potential new liquid biopsy strategy. In addition to dysregulated expression, functional switches that will be detected by analyzing intracellular localization of the molecules should also be perceived as a signal of ncRNA-induced resistance. Basic research is moving from a histological (tissue based) and bulk molecular approach to a single cell approach. We want to point out that this step will not be sufficient for understanding the functions of ncRNAs and a subcellular approach will be necessary. This will be achieved only by performing a single cell spatial non-coding-transcriptomics, which we believe is the next methodological breakthrough that we need. Of note, the location in which miRNAs (the best studied ncRNAs) perform their regulatory function is still a matter of debate.6 Hence, we still need to answer fundamental questions, and we will probably be surprised when we will discover much more mature miRNAs with nuclear localization than expected. Finally, the development of a single cell sequencing technique for ncRNAs will also answer questions regarding ncRNAs’ intratumoral heterogeneity. Currently, single cell transcriptomics techniques can retrieve only poly-adenylated RNAs.166 For example, by using such a method in patients with triple-negative breast cancer and intrinsic resistance to neoadjuvant chemotherapy versus treatment-sensitive patients, Shaath et al. observed that lncRNAs can be used to cluster the patients in these subgroups and that five transcripts of the MALAT1 gene are specifically upregulated in resistant patients.167 This kind of data shows us how much potential there is in moving ncRNA research to the single cell level and ultimately to the subcellular level.
An additional strength of ncRNAs is that these molecules are secreted in virtually all biological fluids. This makes them potential biomarkers, but unfortunately only few ncRNAs have been confirmed for this function. The reason behind is probably, again, heterogeneity, which is also the strength of ncRNAs as potential future biomarkers for personalized medicine. So, this field of research needs to be moved from the diagnostic/screening setting to a sub-classification and response to therapy setting. This type of research needs very well annotated cohorts of patients, which most previous studies lacked. The methodology needs also to be improved, by checking the expression of ncRNAs with specific bio-fluid localization (i.e., bound to proteins, bound to lipids, or intravesicular), the specificity of ncRNA diagnosis can be increased. We envision that, in the near future, ncRNAs could achieve clinical use as biomarkers, most probably in combination with complementary methods—for example, in combination with circulating tumor cells, protein biomarkers, or even metabolites.168
In summary, because ncRNAs are key regulators and predictors of cancer therapy resistance, they could function as therapeutic adjuvants and as components of a tumor-agnostic therapeutic strategy to improve anti-cancer response within existing therapeutic modalities, including chemotherapy, radiotherapy, ICIs, and targeted therapy. However, some key challenges remain and limit the clinical application for ncRNA therapeutics—including issues associated with tolerability, toxicity, and off-target effects, which need to be further elucidated.
References
Dragomir, M. P. et al. FuncPEP: A database of functional peptides encoded by non-coding RNAs. Noncoding RNA 6, 41 (2020).
Carrington, J. C. & Ambros, V. Role of microRNAs in plant and animal development. Science 301, 336–338 (2003).
Pang, K. C., Frith, M. C. & Mattick, J. S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22, 1–5 (2006).
Chen, L. L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17, 205–211 (2016).
Bartel, D. P. Metazoan MicroRNAs. Cell 173, 20–51 (2018).
Dragomir, M. P., Knutsen, E. & Calin, G. A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 38, 379–394 (2021).
Dragomir, M. P., Knutsen, E. & Calin, G. A. SnapShot: unconventional miRNA functions. Cell 174, 1038–1038 (2018).
Kozomara, A. et al. miRBase: from microRNA sequnces to function. Nucleic Acids Res 47, D155–D162 (2019).
Londin, E. et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl Acad. Sci. 112, E1106–E1115 (2015).
Ha, M., Pang, M., Agarwal, V. & Chen, Z. J. Interspecies regulation of microRNAs and their targets. Biochim Biophys. Acta 1779, 735–742 (2008).
Calin, G. A. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 99, 15524–15529 (2002).
Anfossi, S., Babayan, A., Pantel, K. & Calin, G. A. Clinical utility of circulating non-coding RNAs - an update. Nat. Rev. Clin. Oncol. 15, 541–563 (2018).
Lin, S. & Gregory, R. I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 (2015).
Song, S. & Ajani, J. A. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 10, 109–118 (2013).
Hong, D. S. et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 122, 1630–1637 (2020).
Fabbri, M., Girnita, L., Varani, G. & Calin, G. A. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 29, 1377–1388 (2019).
Pisignano, G. et al. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat. Commun. 8, 15622 (2017).
Carlevaro-Fita, J. & Johnson, R. Global positioning system: understanding long noncoding rnas through subcellular localization. Mol. Cell 73, 869–883 (2019).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet 17, 47–62 (2016).
Dragomir, M. P., Kopetz, S., Ajani, J. A. & Calin, G. A. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut 69, 748–763 (2020).
Ji, P. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Chevli, K. K. et al. Urinary PCA3 as a predictor of prostate cancer in a cohort of 3073 men undergoing initial prostate biopsy. J. Urol. 191, 1743–1748 (2014).
Calin, G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007).
Fabris, L. & Calin, G. A. Understanding the genomic ultraconservations: T-UCRs and cancer. Int Rev. Cell Mol. Biol. 333, 159–172 (2017).
Rigoutsos, I. et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 18, 98 (2017).
Pichler, M. et al. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut 69, 1818–1831 (2020).
Rigoutsos, I. et al. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc. Natl Acad. Sci. 103, 6605–6610 (2006).
Dragomir, M. P. et al. The non-coding RNome after splenectomy. J. Cell Mol. Med. 23, 7844–7858 (2019).
Dragomir, M. & Calin, G. A. Circular RNAs in cancer - lessons learned from microRNAs. Front Oncol. 8, 179 (2018).
Ruskin, B. & Green, M. R. An RNA processing activity that debranches RNA lariats. Science 229, 135–140 (1985).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Du, W. W. et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 24, 357–370 (2017).
Salzman, J. et al. Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777 (2013).
Redis, R. S. et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol. Cell 61, 520–534 (2016).
Meyer, M. et al. Profiling the non-genetic origins of cancer drug resistance with a single-cell functional genomics approach using predictive cell dynamics. Cell Syst. 11, 367–374 (2020).
Pardini, B., Sabo, A. A., Birolo, G. & Calin, G. A. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers 11, 1170 (2019).
Fabbri, M. et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. 109, E2110–E2116 (2012).
Dragomir, M. et al. Using microRNA networks to understand cancer. Int. J. Mol. Sci. 19, 1871 (2018).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283 (2016).
Longley, D. B. & Johnston, P. G. Molecular mechanisms of drug resistance. J. Pathol. 205, 275–292 (2005).
Lippert, T. H., Ruoff, H. J. & Volm, M. Intrinsic and acquired drug resistance in malignant tumors. The main reason for therapeutic failure. Arzneimittelforschung 58, 261–264 (2008).
Santoni-Rugiu, E. et al. Intrinsic resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: differences and similarities with acquired resistance. Cancers 11, 923 (2019).
Yu, K. D. et al. Breast cancer patients with estrogen receptor-negative/progesterone receptor-positive tumors: being younger and getting less benefit from adjuvant tamoxifen treatment. J. Cancer Res. Clin. Oncol. 134, 1347–1354 (2008).
Vinogradov, S. & Wei, X. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine. 7, 597–615 (2012).
Chen, Z. et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 370, 153–164 (2016).
Neel, D. S. & Bivona, T. G. Resistance is futile: overcoming resistance to targeted therapies in lung adenocarcinoma. NPJ. Precis. Oncol. 1, 1–6 (2017).
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Ling, H. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013).
Chen, B. et al. The long noncoding RNA CCAT2 induces chromosomal instability through BOP1-AURKB signaling. Gastroenterology 159, 2146–2162 (2020).
He, W. et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 38, 4637–4654 (2019).
Drayton, R. M. et al. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 20, 1990–2000 (2014).
Huang, X. et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 18, 71 (2019).
Ellis, L. M. & Hicklin, D. J. Resistance to targeted therapies: refining anticancer therapy in the era of molecular oncology. Clin. Cancer Res. 15, 7471–7478 (2009).
Molina, A. M. et al. Sunitinib objective response in metastatic renal cell carcinoma: analysis of 1059 patients treated on clinical trials. Eur. J. Cancer 50, 351–358 (2014).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Lu, Y. et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat. Med. 23, 1331–1341 (2017).
Chen, J. et al. Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett. 367, 1–11 (2015).
Xu, W. P. et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut 69, 1309–1321 (2020).
Xu, J. et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target Ther. 5, 298 (2020).
Park, S. Y., Kim, J. Y., Jun, Y. & Nam, J. S. Strategies to tackle radiation resistance by penetrating cancer stem cell line of scrimmage. Recent Pat. Anticancer Drug Disco. 13, 18–39 (2018).
Barker, H. E., Paget, J. T., Khan, A. A. & Harrington, K. J. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat. Rev. Cancer 15, 409–425 (2015).
Guo, Z. et al. LncRNA linc00312 suppresses radiotherapy resistance by targeting DNA-PKcs and impairing DNA damage repair in nasopharyngeal carcinoma. Cell Death Dis. 12, 69 (2021).
Pajic, M. et al. miR-139-5p modulates radiotherapy resistance in breast cancer by repressing multiple gene networks of DNA repair and ROS defense. Cancer Res. 78, 501–515 (2018).
Yuan, Y. et al. miR-410 induces both epithelial-mesenchymal transition and radioresistance through activation of the PI3K/mTOR pathway in non-small cell lung cancer. Signal Transduct. Target Ther. 5, 85 (2020).
Xia, X. et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer 18, 131 (2019).
Schadendorf, D. et al. pooled analysis of long-term survival data from phase IIand phase III trials of ipilimumab in unresectable or metastatic melanoma. J.Clin. Oncol 33, 1889–1894 (2015).
Jenkins, R. W., Barbie, D. A. & Flaherty, K. T. Mechanisms of resistance to immune checkpoint inhibitors. Br. J. Cancer 118, 9–16 (2018).
Hu, Q. et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 20, 835–851 (2019).
Gebhardt, C. et al. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with ipilimumab. Clin. Cancer Res. 21, 5453–5459 (2015).
Huber, V. et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Invest 128, 5505–5516 (2018).
Huang, X. Y. et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 19, 92 (2020).
Zhang, P. F. et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19, 110 (2020).
Mayeux, R. Biomarkers: potential uses and limitations. NeuroRx 1, 182–188 (2004).
Henry, N. L. & Hayes, D. F. Cancer biomarkers. Mol. Oncol. 6, 140–146 (2012).
Aberuyi, N., Rahgozar, S., Ghodousi, E. S. & Ghaedi, K. Drug resistance biomarkers and their clinical applications in childhood acute lymphoblastic leukemia. Front Oncol. 9, 1496 (2019).
Bolha, L., Ravnik-Glavac, M. & Glavac, D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers 2017, 7243968 (2017).
Lan, H., Lu, H., Wang, X. & Jin, H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed. Res. Int. 2015, 125094 (2015).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Hu, G. et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 14, 1190–1198 (2013).
Zhang, Y. et al. The lncRNA Snhg1-Vps13D vesicle trafficking system promotes memory CD8 T cell establishment via regulating the dual effects of IL-7 signaling. Signal Transduct. Target Ther. 6, 126 (2021).
Alix-Panabieres, C. The future of liquid biopsy. Nature 579, S9 (2020).
Barth, D. A. et al. Circulating non-coding RNAs in renal cell carcinoma-pathogenesis and potential implications as clinical biomarkers. Front Cell Dev. Biol. 8, 828 (2020).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. 108, 5003–5008 (2011).
Rayner, K. J. & Hennessy, E. J. Extracellular communication via microRNA: lipid particles have a new message. J. Lipid Res. 54, 1174–1181 (2013).
Vickers, K. C. & Remaley, A. T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipido. 23, 91–97 (2012).
Zhang, H. G. & Grizzle, W. E. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am. J. Pathol. 184, 28–41 (2014).
Dragomir, M., Chen, B. & Calin, G. A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 7, S243–S252 (2018).
Zhang, J. et al. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs 25, 346–352 (2014).
Kjersem, J. B. et al. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 8, 59–67 (2014).
Rasmussen, M. H. et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol. Oncol. 7, 637–646 (2013).
Rasmussen, M. H. et al. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat. Commun. 7, 12436 (2016).
Xu, W. et al. MiR-145 in cancer therapy resistance and sensitivity: A comprehensive review. Cancer Sci. 111, 3122–3131 (2020).
Pan, Y. J., Zhuang, Y., Zheng, J. N. & Pei, D. S. MiR-106a: Promising biomarker for cancer. Bioorg. Med. Chem. Lett. 26, 5373–5377 (2016).
Lu, H. et al. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy. Biosci. Rep. 39, 5 (2019).
Zhu, W. et al. Dynamics of circulating microRNAs as a novel indicator of clinical response to neoadjuvant chemotherapy in breast cancer. Cancer Med. 7, 4420–4433 (2018).
Xiong, Y. et al. iASPP induces EMT and cisplatin resistance in human cervical cancer through miR-20a-FBXL5/BTG3 signaling. J. Exp. Clin. Cancer Res. 36, 48 (2017).
Zhang, Y. et al. MiR-20a induces cell radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 92, 1132–1140 (2015).
Pu, Y. et al. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int. 16, 64 (2016).
Garcia-Garcia, F. et al. miR1455p is associated with pathological complete response to neoadjuvant chemotherapy and impairs cell proliferation by targeting TGFbetaR2 in breast cancer. Oncol. Rep. 41, 3527–3534 (2019).
McGuire, A. et al. Prospective assessment of systemic MicroRNAs as markers of response to neoadjuvant chemotherapy in breast cancer. Cancers 12, 1820 (2020).
Lim, E. L. et al. MicroRNA expression-based model indicates event-free survival in pediatric acute myeloid leukemia. J. Clin. Oncol. 35, 3964–3977 (2017).
Jin, L. et al. Serum microRNAs as potential new biomarkers for cisplatin resistance in gastric cancer patients. PeerJ 8, e8943 (2020).
Qi, M., Liu, D. & Zhang, S. MicroRNA-21 contributes to the discrimination of chemoresistance in metastatic gastric cancer. Cancer Biomark. 18, 451–458 (2017).
Hamano, R. et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 17, 3029–3038 (2011).
Xiao, Y., Yurievich, U. A. & Yosypovych, S. V. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget 8, 83171–83182 (2017).
Li, L. et al. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol. Rep. 38, 1383–1392 (2017).
Zeng, Y. et al. LncRNA profile study reveals a three-LncRNA signature associated with the pathological complete response following neoadjuvant chemotherapy in breast cancer. Front Pharm. 10, 574 (2019).
Liu, R. et al. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Scientific Rep. 7, 1–10 (2017).
Wang, S. et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271 (2008).
Hansen, T. F. et al. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br. J. Cancer 112, 624–629 (2015).
Tan, W. et al. miR-126-3p contributes to sorafenib resistance in hepatocellular carcinoma via downregulating SPRED1. Ann. Transl. Med. 9, 38 (2021).
Lechman, E. R. et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 29, 214–228 (2016).
Caporali, S. et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J. Exp. Clin. Cancer Res. 38, 272 (2019).
Hoppe, R. et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur. J. Cancer 49, 3598–3608 (2013).
Rinnerthaler, G. et al. Low expression of miR-20a-5p predicts benefit to bevacizumab in metastatic breast cancer patients treated within the TANIA Phase III trial. J. Clin. Med. 9, 1663 (2020).
Zhao, Q. et al. Circulating miRNAs is a potential marker for gefitinib sensitivity and correlation with EGFR mutational status in human lung cancers. Am. J. Cancer Res. 5, 1692–1705 (2015).
Du, W. et al. The miR6253p/AXL axis induces nonT790M acquired resistance to EGFRTKI via activation of the TGFbeta/Smad pathway and EMT in EGFRmutant nonsmall cell lung cancer. Oncol. Rep. 44, 185–195 (2020).
Kou, Y., Yang, R. & Wang, Q. Serum miR-518e-5p is a potential biomarker for secondary imatinib-resistant gastrointestinal stromal tumor. J. Biosci. 43, 1015–1023 (2018).
Bai, Y. & Su, X. Updates to the drug-resistant mechanism of proteasome inhibitors in multiple myeloma. Asia Pac. J. Clin. Oncol. 17, 29–35 (2021).
Robak, P. et al. The value of serum MicroRNA expression signature in predicting refractoriness to bortezomib-based therapy in multiple myeloma patients. Cancers 12, 2569 (2020).
Jin, J. et al. Methylation-associated silencing of miR-193b improves the radiotherapy sensitivity of esophageal cancer cells by targeting cyclin D1 in areas with zinc deficiency. Radiother. Oncol. 150, 104–113 (2020).
Bulibu, J. et al. Association between polymorphisms in the promoter region of microRNA-34b/c and the chemoradiotherapy efficacy for locally advanced esophageal squamous cell carcinoma in chinese han population. Pathol. Oncol. Res. 25, 421–427 (2017).
Ferrando, L. et al. Development of a long non-coding RNA signature for prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal adenocarcinoma. PLoS One 15, e0226595 (2020).
Dragomir, M., Chen, B., Fu, X. & Calin, G. A. Key questions about the checkpoint blockade-are microRNAs an answer? Cancer Biol. Med. 15, 103–115 (2018).
Smolle, M. A., Calin, H. N., Pichler, M. & Calin, G. A. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 284, 1952–1966 (2017).
Grenda, A. & Krawczyk, P. New dancing couple: PD-L1 and MicroRNA. Scand. J. Immunol. 86, 130–134 (2017).
Schrock, A. B. et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 30, 1096–1103 (2019).
Xia, Y. et al. miRNA-based feature classifier is associated with tumor mutational burden in head and neck squamous cell carcinoma. Biomed. Res. Int. 2020, 1686480 (2020).
Zhou, H. et al. An miRNA signature associated with tumor mutation burden in endometrial cancer. Biosci. Rep. 40, BSR20203398 (2020).
Lv, Y. et al. MiRNA expression patterns are associated with tumor mutational burden in lung adenocarcinoma. Oncoimmunology 8, e1629260 (2019).
Ding, C. et al. Exploration of the associations of lncRNA expression patterns with tumor mutation burden and prognosis in colon cancer. Onco. Targets Ther. 14, 2893–2909 (2021).
Yang, D. et al. Long non-coding rna expression patterns in stomach adenocarcinoma serve as an indicator of tumor mutation burden and are associated with tumor-infiltrating lymphocytes and microsatellite instability. Front Cell Dev. Biol. 9, 618313 (2021).
Sudo, K. et al. Identification of serum microRNAs predicting the response of esophageal squamous-cell carcinoma to nivolumab. Jpn J. Clin. Oncol. 50, 114–121 (2020).
Shukuya, T. et al. Circulating MicroRNAs and extracellular vesicle-containing MicroRNAs as response biomarkers of anti-programmed cell death protein 1 or programmed death-ligand 1 therapy in NSCLC. J. Thorac. Oncol. 15, 1773–1781 (2020).
Omar, H. A. et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 286, 3540–3557 (2019).
Mittelbrunn, M. et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282 (2011).
Winkle, M. & El-Daly, S. M. Fabbÿri, M. & Calin, G. A. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 20, 629–651 (2021).
Crooke, S. T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 27, 70–77 (2017).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689 (2005).
Ottosen, S. et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 59, 599–608 (2015).
Zhou, T., Kim, Y. & MacLeod, A. R. Targeting long noncoding rna with antisense oligonucleotide technology as cancer therapeutics. Methods Mol. Biol. 1402, 199–213 (2016).
Dana, H. et al. Molecular mechanisms and biological functions of siRNA. Int J. Biomed. Sci. 13, 48–57 (2017).
Moore, C. B., Guthrie, E. H., Huang, M. T. & Taxman, D. J. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol. Biol. 629, 141–158 (2010).
Yang, J. et al. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 15, 35–43 (2018).
Ebert, M. S. & Sharp, P. A. MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050 (2010).
Wen, D., Danquah, M., Chaudhary, A. K. & Mahato, R. I. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J. Control Release 219, 237–247 (2015).
Pedram Fatemi, R. et al. Screening for small-molecule modulators of long noncoding RNA-protein interactions using alphascreen. J. Biomol. Screen 20, 1132–1141 (2015).
Bader, A. G., Brown, D. & Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 70, 7027–7030 (2010).
Lehmann, S. M. et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835 (2012).
LaCasse, E. C. Pulling the plug on a cancer cell by eliminating XIAP with AEG35156. Cancer Lett. 332, 215–224 (2013).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Disco. 19, 673–694 (2020).
Jia, Z. et al. pPB Peptide-Mediated siRNA-loaded stable nucleic acid lipid nanoparticles on targeting therapy of hepatic fibrosis. Mol. Pharm. 15, 53–62 (2018).
Akbarzadeh, A. et al. Liposome: classification, preparation, and applications. Nanoscale Res. Lett. 8, 102 (2013).
Kristen, A. V. et al. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 9, 5–23 (2019).
MacEwan, S. R., Callahan, D. J. & Chilkoti, A. Stimulus-responsive macromolecules and nanoparticles for cancer drug delivery. Nanomedicine 5, 793–806 (2010).
Wang, X. et al. MiRNA delivery system based on stimuli-responsive gold nanoparticle aggregates for multimodal tumor therapy. ACS Appl. Bio. Mater. 2, 2833–2839 (2019).
Ekin, A., Karatas, O. F., Culha, M. & Ozen, M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J. Gene Med. 16, 331–335 (2014).
Springer, A. D. & Dowdy, S. F. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28, 109–118 (2018).
Balwani, M. et al. Phase 3 Trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Engl. J. Med. 382, 2289–2301 (2020).
Dugal-Tessier, J., Thirumalairajan, S. & Jain, N. Antibody-oligonucleotide conjugates: a twist to antibody-drug conjugates. J. Clin. Med. 10, 838 (2021).
Kordelas, L. et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 28, 970–973 (2014).
Offin, M., Liu, D. & Drilon, A. Tumor-agnostic drug development. Am. Soc. Clin. Oncol. Educ. Book 38, 184–187 (2018).
Marcus, L., Lemery, S. J., Keegan, P. & Pazdur, R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin. Cancer Res. 25, 3753–3758 (2019).
Drilon, A. et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol. 21, 261–270 (2020).
Alessio, E. et al. A single cell but many different transcripts: a journey into the world of long non-coding RNAs. Int. J. Mol. Sci. 21, 302 (2020).
Shaath, H. et al. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Disco. 7, 23 (2021).
Moisoiu, V. et al. SERS liquid biopsy: An emerging tool for medical diagnosis. Colloids Surf. B Biointerfaces 208, 112064 (2021).
Pihan, G. A. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front Oncol. 3, 277 (2013).
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 15, 139–150 (2018).
Shah, M. Y. et al. Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations. Genome Res. 28, 432–447 (2018).
Lee, S. et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016).
Xu, C. et al. Comparative safety of immune checkpoint inhibitors in cancer: systematic review and network meta-analysis. BMJ 363, k4226 (2018).
Wei, J. et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro. Oncol. 18, 639–648 (2016).
Zhang, X. L., Xu, L. L. & Wang, F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 41, 1056–1064 (2017).
Zhen, S. et al. Synergistic antitumor effect on bladder cancer by rational combination of programmed cell death 1 blockade and CRISPR-Cas9-mediated long non-coding RNA urothelial carcinoma associated 1 knockout. Hum. Gene Ther. 29, 1352–1363 (2018).
Acknowledgements
We thank Bryan Tutt, Scientific Editor, Research Medical Library, MDACC for editorial support. The work of B.C. is supported by National Natural Science Foundation of China (No. 81902462). M.P.D. is a participant in the BIH-Charité Junior Clinical Scientist Program funded by the Charité – Universitätsmedizin Berlin and the Berlin Institute of Health. G.A.C. is the Felix L. Haas Endowed Professor in Basic Science. Work in G.A.C.’s laboratory is supported by NCI grants 1R01 CA182905-01 and 1R01CA222007-01A1, NIGMS grant 1R01GM122775-01, DoD Idea Award W81XWH2110030, a Team DOD grant in Gastric Cancer, a Chronic Lymphocytic Leukemia Moonshot Flagship project, a CLL Global Research Foundation 2019 grant, a CLL Global Research Foundation 2020 grant, The G. Harold & Leila Y. Mathers Foundation, a grant from Torrey Coast Foundation, and an Institutional Research Grant and Development Grant associated with the Brain SPORE 2P50CA127001.
Author information
Authors and Affiliations
Contributions
G.A.C., B.C., and M.P.D. provided direction and instruction in preparing this manuscript. G.A.C., B.C., M.P.D., Y.C., and Q.L., D.H. reviewed the literature and drafted this manuscript. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
G.A.C. is the scientific founder of Ithax Pharmaceuticals. All other authors declare no conflict of interest. G.A.C. is the editorial board member of Signal Transduction and Targeted Therapy, but he has not been involved in the process of the manuscript handling.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, B., Dragomir, M.P., Yang, C. et al. Targeting non-coding RNAs to overcome cancer therapy resistance. Sig Transduct Target Ther 7, 121 (2022). https://doi.org/10.1038/s41392-022-00975-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-022-00975-3
This article is cited by
-
KIT mutations and expression: current knowledge and new insights for overcoming IM resistance in GIST
Cell Communication and Signaling (2024)
-
TGF-β-activated circRYK drives glioblastoma progression by increasing VLDLR mRNA expression and stability in a ceRNA- and RBP-dependent manner
Journal of Experimental & Clinical Cancer Research (2024)
-
Non-coding RNAs as potential therapeutic targets for receptor tyrosine kinase signaling in solid tumors: current status and future directions
Cancer Cell International (2024)
-
Ascites exosomal lncRNA PLADE enhances platinum sensitivity by inducing R-loops in ovarian cancer
Oncogene (2024)
-
FAK-LINC01089 negative regulatory loop controls chemoresistance and progression of small cell lung cancer
Oncogene (2024)