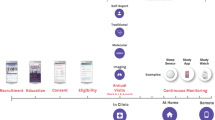
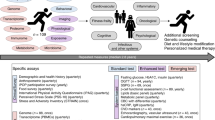
Abstract
Modern health care faces several serious challenges, including an ageing population and its inherent burden of chronic diseases, rising costs and marginal quality metrics. By assessing and optimizing the health trajectory of each individual using a data-driven personalized approach that reflects their genetics, behaviour and environment, we can start to address these challenges. This assessment includes longitudinal phenome measures, such as the blood proteome and metabolome, gut microbiome composition and function, and lifestyle and behaviour through wearables and questionnaires. Here, we review ongoing large-scale genomics and longitudinal phenomics efforts and the powerful insights they provide into wellness. We describe our vision for the transformation of the current health care from disease-oriented to data-driven, wellness-oriented and personalized population health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
WHO Department of Data and Analytics, Division of Data, Analytics and Delivery for Impact. WHO Methods and Data Sources for Life Tables 1990–2019. https://www.who.int/docs/default-source/gho-documents/global-health-estimates/ghe2019_life-table-methods.pdf (2020).
Hajat, C. & Stein, E. The global burden of multiple chronic conditions: a narrative review. Prev. Med. Rep. 12, 284–293 (2018).
Schork, N. J. Personalized medicine: time for one-person trials. Nature 520, 609–611 (2015).
Senn, S. Statistical pitfalls of personalized medicine. Nature 563, 619–621 (2018).
Dunlap, N. E. et al. Observations from the field: reporting quality metrics in health care. NAM Perspect. https://doi.org/10.31478/201607e (2016).
AHRQ. 2022 National Healthcare Quality and Disparities Report. Agency for Healthcare Research and Quality https://www.ahrq.gov/research/findings/nhqrdr/nhqdr22/index.html (2023).
Yurkovich, J. T. & Hood, L. Blood is a window into health and disease. Clin. Chem. 65, 1204–1206 (2019).
Mooradian, A. D. The merits and the pitfalls of low carbohydrate diet: a concise review. J. Nutr. Health Aging 24, 805–808 (2020).
Lee, P. et al. Digital health COVID-19 impact assessment: lessons learned and compelling needs. NAM Perspect. https://doi.org/10.31478/202201c (2022).
Pennisi, E. A $100 genome? New DNA sequencers could be a ‘game changer’ for biology, medicine. Science https://doi.org/10.1126/science.add5060 (2022).
Bowcock, A. M. Genomics: guilt by association. Nature 447, 645–646 (2007).
Kahl, V. F. S. et al. Telomere length measurement by molecular combing. Front. Cell Dev. Biol. 8, 493 (2020).
Reinius, L. E. et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7, e41361 (2012).
Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023).
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 (2009).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Yousefi, P. D. et al. DNA methylation-based predictors of health: applications and statistical considerations. Nat. Rev. Genet. 23, 369–383 (2022).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018).
Lio, C.-W. J., Yuita, H. & Rao, A. Dysregulation of the TET family of epigenetic regulators in lymphoid and myeloid malignancies. Blood 134, 1487–1497 (2019).
Guo, M., Peng, Y., Gao, A., Du, C. & Herman, J. G. Epigenetic heterogeneity in cancer. Biomark. Res. 7, 23 (2019).
Apicella, C., Ruano, C. S. M., Méhats, C., Miralles, F. & Vaiman, D. The role of epigenetics in placental development and the etiology of preeclampsia. Int. J. Mol. Sci. 20, 2837 (2019).
Lim, U. & Song, M.-A. Dietary and lifestyle factors of DNA methylation. Methods Mol. Biol. 863, 359–376 (2012).
Rozek, L. S., Dolinoy, D. C., Sartor, M. A. & Omenn, G. S. Epigenetics: relevance and implications for public health. Annu. Rev. Public Health 35, 105–122 (2014).
Kim, C. H. et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep. 8, 8382 (2018).
Petrera, A. et al. Multiplatform approach for plasma proteomics: complementarity of Olink Proximity Extension Assay technology to mass spectrometry-based protein profiling. J. Proteome Res. 20, 751–762 (2021).
Eldjarn, G. H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023).
Chandramouli, K. & Qian, P.-Y. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum. Genom. Proteom. 2009, 239204 (2009).
Güntner, A. T. et al. Breath sensors for health monitoring. ACS Sens. 4, 268–280 (2019).
Kennedy, A. D. et al. Metabolomics in the clinic: a review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 53, 1143–1154 (2018).
Würtz, P. et al. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am. J. Epidemiol. 186, 1084–1096 (2017).
Tasoglu, S. Toilet-based continuous health monitoring using urine. Nat. Rev. Urol. 19, 219–230 (2022).
Pace, N. R. A molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997).
Heinken, A. et al. Genome-scale metabolic reconstruction of 7,302 human microorganisms for personalized medicine. Nat. Biotechnol. 41, 1320–1331 (2023).
Mayer, E. A., Nance, K. & Chen, S. The gut–brain axis. Annu. Rev. Med. 73, 439–453 (2022).
Tilg, H., Adolph, T. E. & Trauner, M. Gut–liver axis: pathophysiological concepts and clinical implications. Cell Metab. 34, 1700–1718 (2022).
Bosco, N. & Noti, M. The aging gut microbiome and its impact on host immunity. Genes Immun. 22, 289–303 (2021).
Andoh, A. & Nishida, A. Alteration of the gut microbiome in inflammatory bowel disease. Digestion 104, 16–23 (2023).
Nichols, R. G., Peters, J. M. & Patterson, A. D. Interplay between the host, the human microbiome, and drug metabolism. Hum. Genom. 13, 27 (2019).
Wilmanski, T. et al. Heterogeneity in statin responses explained by variation in the human gut microbiome. Med 3, 388–405.e6 (2022).
Wilmanski, T., Rappaport, N., Diener, C., Gibbons, S. M. & Price, N. D. From taxonomy to metabolic output: what factors define gut microbiome health? Gut Microbes 13, 1–20 (2021).
Abdill, R. J., Adamowicz, E. M. & Blekhman, R. Public human microbiome data are dominated by highly developed countries. PLoS Biol. 20, e3001536 (2022).
Ip, J. E. Wearable devices for cardiac rhythm diagnosis and management. JAMA 321, 337–338 (2019).
Ates, H. C. et al. End-to-end design of wearable sensors. Nat. Rev. Mater. 7, 887–907 (2022).
Lee, I., Probst, D., Klonoff, D. & Sode, K. Continuous glucose monitoring systems — current status and future perspectives of the flagship technologies in biosensor research. Biosens. Bioelectron. 181, 113054 (2021).
Taj, F., Klein, M. C. A. & van Halteren, A. Digital health behavior change technology: bibliometric and scoping review of two decades of research. JMIR mHealth uHealth 7, e13311 (2019).
Öhman, F., Hassenstab, J., Berron, D., Schöll, M. & Papp, K. V. Current advances in digital cognitive assessment for preclinical Alzheimer’s disease. Alzheimers Dement. 13, e12217 (2021).
Nahum, M., Lee, H. & Merzenich, M. M. Principles of neuroplasticity-based rehabilitation. Prog. Brain Res. 207, 141–171 (2013).
Lindner, N., Kuwabara, A. & Holt, T. Non-invasive and minimally invasive glucose monitoring devices: a systematic review and meta-analysis on diagnostic accuracy of hypoglycaemia detection. Syst. Rev. 10, 145 (2021).
Andreou, C., Weissleder, R. & Kircher, M. F. Multiplexed imaging in oncology. Nat. Biomed. Eng. 6, 527–540 (2022).
Hu, H. et al. A wearable cardiac ultrasound imager. Nature 613, 667–675 (2023).
Williams, S. A. et al. Plasma protein patterns as comprehensive indicators of health. Nat. Med. 25, 1851–1857 (2019).
Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit. Health 1, e271–e297 (2019).
Wong, B. L. H. et al. The dawn of digital public health in Europe: implications for public health policy and practice. Lancet Reg. Health Eur. 14, 100316 (2022).
Boyd, A. D. et al. Potential bias and lack of generalizability in electronic health record data: reflections on health equity from the National Institutes of Health Pragmatic Trials Collaboratory. J. Am. Med. Inform. Assoc. 30, 1561–1566 (2023).
Joshua Lin, K. et al. Longitudinal data discontinuity in electronic health records and consequences for medication effectiveness studies. Clin. Pharmacol. Ther. 111, 243–251 (2022).
Carey, D. J. et al. The Geisinger MyCode Community Health Initiative: an electronic health record-linked biobank for precision medicine research. Genet. Med. 18, 906–913 (2016).
Vatsalan, D., Christen, P. & Verykios, V. S. A taxonomy of privacy-preserving record linkage techniques. Inf. Syst. 38, 946–969 (2013).
Jain, A. & Srivastava, N. Privacy-preserving record linkage with block-chains. In Proc. Cyber Security, Privacy and Networking 61–70 (Springer Nature, 2022).
Kumar, M. & Mostafa, J. Research evidence on strategies enabling integration of electronic health records in the health care systems of low- and middle-income countries: a literature review. Int. J. Health Plann. Manag. 34, e1016–e1025 (2019).
Ebrahim, A. et al. Do genome-scale models need exact solvers or clearer standards? Mol. Syst. Biol. 11, 831 (2015).
Deutsch, E. W. et al. Proteomics Standards Initiative at twenty years: current activities and future work. J. Proteome Res. 22, 287–301 (2023).
Koistinen, V. et al. Towards a Rosetta stone for metabolomics: recommendations to overcome inconsistent metabolite nomenclature. Nat. Metab. 5, 351–354 (2023).
Carey, M. A., Dräger, A., Beber, M. E., Papin, J. A. & Yurkovich, J. T. Community standards to facilitate development and address challenges in metabolic modeling. Mol. Syst. Biol. 16, e9235 (2020).
Köhler, S. et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 49, D1207–D1217 (2021).
Piekos, S. N. et al. Biomedical Data Commons (BMDC) prioritizes B-lymphocyte non-coding genetic variants in type 1 diabetes. PLoS Comput. Biol. 17, e1009382 (2021).
Fecho, K. et al. Progress toward a universal biomedical data translator. Clin. Transl Sci. 15, 1838–1847 (2022).
Su, Y. et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 183, 1479–1495.e20 (2020).
Orth, M. F. et al. Systematic multi-omics cell line profiling uncovers principles of Ewing sarcoma fusion oncogene-mediated gene regulation. Cell Rep. 41, 111761 (2022).
Price, N. D. et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 35, 747–756 (2017).
Wilmanski, T. et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat. Biotechnol. 37, 1217–1228 (2019).
Magis, A. T. et al. Untargeted longitudinal analysis of a wellness cohort identifies markers of metastatic cancer years prior to diagnosis. Sci. Rep. 10, 16275 (2020).
Earls, J. C. et al. Multi-omic biological age estimation and its correlation with wellness and disease phenotypes: a longitudinal study of 3,558 individuals. J. Gerontol. A Biol. Sci. Med. Sci. 74, S52–S60 (2019).
Watanabe, K. et al. Multiomic signatures of body mass index identify heterogeneous health phenotypes and responses to a lifestyle intervention. Nat. Med. 29, 996–1008 (2023).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23, 715–727 (2022).
Shah, N. R. & Braverman, E. R. Measuring adiposity in patients: the utility of body mass index (BMI), percent body fat, and leptin. PLoS ONE 7, e33308 (2012).
Tomiyama, A. J., Hunger, J. M., Nguyen-Cuu, J. & Wells, C. Misclassification of cardiometabolic health when using body mass index categories in NHANES 2005-2012. Int. J. Obes. 40, 883–886 (2016).
Franzosa, E. A. et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305 (2019).
Xavier, J. B. et al. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer Res. 6, 192–204 (2020).
Girinathan, B. P. et al. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e7 (2021).
Wu, H. et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 32, 379–390.e3 (2020).
Johnson, J. P. et al. Generally-healthy individuals with aberrant bowel movement frequencies show enrichment for microbially-derived blood metabolites associated with impaired kidney function. Preprint at bioRxiv https://doi.org/10.1101/2023.03.04.531100 (2023).
Bohmann, N. et al. Microbial community-scale metabolic modeling predicts personalized short-chain-fatty-acid production profiles in the human gut. Preprint at bioRxiv https://doi.org/10.1101/2023.02.28.530516 (2023).
Sharp, S. A., Weedon, M. N., Hagopian, W. A. & Oram, R. A. Clinical and research uses of genetic risk scores in type 1 diabetes. Curr. Opin. Genet. Dev. 50, 96–102 (2018).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Visscher, P. M., Yengo, L., Cox, N. J. & Wray, N. R. Discovery and implications of polygenicity of common diseases. Science 373, 1468–1473 (2021).
Marston, N. A. et al. Predictive utility of a coronary artery disease polygenic risk score in primary prevention. JAMA Cardiol. 8, 130–137 (2023).
Wainberg, M. et al. Multiomic blood correlates of genetic risk identify presymptomatic disease alterations. Proc. Natl Acad. Sci. USA 117, 21813–21820 (2020).
Nguengang Wakap, S. et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur. J. Hum. Genet. 28, 165–173 (2020).
Yang, G. et al. The national economic burden of rare disease in the United States in 2019. Orphanet J. Rare Dis. 17, 163 (2022).
Willmen, T. et al. Health economic benefits through the use of diagnostic support systems and expert knowledge. BMC Health Serv. Res. 21, 947 (2021).
Schuermans, N. et al. Shortcutting the diagnostic odyssey: the multidisciplinary Program for Undiagnosed Rare Diseases in adults (UD-PrOZA). Orphanet J. Rare Dis. 17, 210 (2022).
Dhindsa, R. S. et al. Rare variant associations with plasma protein levels in the UK Biobank. Nature 622, 339–347 (2023).
Kerr, K. et al. A scoping review and proposed workflow for multi-omic rare disease research. Orphanet J. Rare Dis. 15, 107 (2020).
Unni, D. R. et al. Biolink model: a universal schema for knowledge graphs in clinical, biomedical, and translational science. Clin. Transl Sci. 15, 1848–1855 (2022).
de Vries, B. M. et al. Explainable artificial intelligence (XAI) in radiology and nuclear medicine: a literature review. Front. Med. 10, 1180773 (2023).
Alloghani, M. et al. in Communications in Computer and Information Science 248–261 (Springer, 2020).
Goh, K.-I. & Choi, I.-G. Exploring the human diseasome: the human disease network. Brief. Funct. Genom. 11, 533–542 (2012).
Xu, H. et al. APRILE: exploring the molecular mechanisms of drug side effects with explainable graph neural networks. Preprint at bioRxiv https://doi.org/10.1101/2021.07.02.450937 (2021).
Ruiz, C., Zitnik, M. & Leskovec, J. Identification of disease treatment mechanisms through the multiscale interactome. Nat. Commun. 12, 1796 (2021).
Qiu, W., Chen, H., Kaeberlein, M. & Lee, S.-I. ExplaiNAble BioLogical Age (ENABL Age): an artificial intelligence framework for interpretable biological age. Lancet Healthy Longev. 4, e711–723 (2023).
van der Velden, B. H. M., Kuijf, H. J., Gilhuijs, K. G. A. & Viergever, M. A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 79, 102470 (2022).
Molani, S. et al. Risk factors for severe COVID-19 differ by age for hospitalized adults. Sci. Rep. 12, 6568 (2022).
Yurkovich, J. T., Tian, Q., Price, N. D. & Hood, L. A systems approach to clinical oncology uses deep phenotyping to deliver personalized care. Nat. Rev. Clin. Oncol. 17, 183–194 (2019).
Hernandez-Boussard, T. et al. Digital twins for predictive oncology will be a paradigm shift for precision cancer care. Nat. Med. 27, 2065–2066 (2021).
Coorey, G., Figtree, G. A., Fletcher, D. F. & Redfern, J. The health digital twin: advancing precision cardiovascular medicine. Nat. Rev. Cardiol. 18, 803–804 (2021).
Popa, E. O., van Hilten, M., Oosterkamp, E. & Bogaardt, M.-J. The use of digital twins in healthcare: socio-ethical benefits and socio-ethical risks. Life Sci. Soc. Policy 17, 6 (2021).
Smarr, B. L. et al. Feasibility of continuous fever monitoring using wearable devices. Sci. Rep. 10, 21640 (2020).
Hua, H. et al. A wipe-based stool collection and preservation kit for microbiome community profiling. Front. Immunol. 13, 889702 (2022).
Meydan, C. et al. Improved gastrointestinal health for irritable bowel syndrome with metagenome-guided interventions. Precis. Clin. Med. 3, 136–146 (2020).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Koulman, A. et al. The development, validation and application of remote blood sample collection in telehealth programmes. J. Telemed. Telecare https://doi.org/10.1177/1357633X221093434 (2022).
Johnson, R. et al. Volumetric absorptive microsampling–LC–MS/MS assays for quantitation of giredestrant in dried human whole blood. Bioanalysis 14, 1377–1389 (2022).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Jiang, L. Y. et al. Health system-scale language models are all-purpose prediction engines. Nature 619, 357–362 (2023).
Mündler, N., He, J., Jenko, S. & Vechev, M. Self-contradictory hallucinations of large language models: evaluation, detection and mitigation. Preprint at arXiv https://doi.org/10.48550/ARXIV.2305.15852 (2023).
Meskó, B. & Topol, E. J. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit. Med. 6, 120 (2023).
Sallam, M. ChatGPT utility in healthcare education, research, and practice: systematic review on the promising perspectives and valid concerns. Healthcare 11, 887 (2023).
Zeggini, E., Gloyn, A. L., Barton, A. C. & Wain, L. V. Translational genomics and precision medicine: moving from the lab to the clinic. Science 365, 1409–1413 (2019).
Oliveira, K. C. S. et al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Mol. Cancer Res. 18, 517–528 (2020).
Cammarota, G. et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 635–648 (2020).
Green, R. C. et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15, 565–574 (2013).
ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet. Med. 17, 68–69 (2015).
Sadee, W., Wang, D., Hartmann, K. & Toland, A. E. Pharmacogenomics: driving personalized medicine. Pharmacol. Rev. 75, 789–814 (2023).
Katsanis, N. et al. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science 293, 2256–2259 (2001).
Alkuraya, F. S. The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Hum. Genet. 132, 1197–1211 (2013).
Sidransky, E. Heterozygosity for a Mendelian disorder as a risk factor for complex disease. Clin. Genet. 70, 275–282 (2006).
Sellami, M., Elrayess, M. A., Puce, L. & Bragazzi, N. L. Molecular big data in sports sciences: state-of-art and future prospects of OMICS-based sports sciences. Front. Mol. Biosci. 8, 815410 (2021).
Dashti, H. S. & Ordovás, J. M. Genetics of sleep and insights into its relationship with obesity. Annu. Rev. Nutr. 41, 223–252 (2021).
Lazaridis, L. et al. Precision neuro-oncology: a pilot analysis of personalized treatment in recurrent glioma. J. Cancer Res. Clin. Oncol. 149, 3513–3526 (2023).
Fatumo, S. et al. A roadmap to increase diversity in genomic studies. Nat. Med. 28, 243–250 (2022).
Farrer, L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278, 1349–1356 (1997).
Rajabli, F. et al. Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 14, e1007791 (2018).
Kulminski, A. M. et al. APOE region molecular signatures of Alzheimer’s disease across races/ethnicities. Neurobiol. Aging 87, 141.e1–141.e8 (2020).
Shah, A. & Kanaya, A. M. Diabetes and associated complications in the South Asian population. Curr. Cardiol. Rep. 16, 476 (2014).
Hills, A. P. et al. Epidemiology and determinants of type 2 diabetes in South Asia. Lancet Diabetes Endocrinol. 6, 966–978 (2018).
Ahmad, S., Fatima, S. S., Rukh, G. & Smith, C. E. Gene lifestyle interactions with relation to obesity, cardiometabolic, and cardiovascular traits among South Asians. Front. Endocrinol. 10, 221 (2019).
Hodgson, S. et al. Integrating polygenic risk scores in the prediction of type 2 diabetes risk and subtypes in British Pakistanis and Bangladeshis: a population-based cohort study. PLoS Med. 19, e1003981 (2022).
Yusuf, S., Reddy, S., Ounpuu, S. & Anand, S. Global burden of cardiovascular diseases: part II: variations in cardiovascular disease by specific ethnic groups and geographic regions and prevention strategies. Circulation 104, 2855–2864 (2001).
Shevchenko, Y. & Bale, S. Clinical versus research sequencing. Cold Spring Harb. Perspect. Med. 6, a025809 (2016).
Rockowitz, S. et al. Children’s rare disease cohorts: an integrative research and clinical genomics initiative. NPJ Genom. Med. 5, 29 (2020).
Schaibley, V. M. et al. Limited genomics training among physicians remains a barrier to genomics-based implementation of precision medicine. Front. Med. 9, 757212 (2022).
Nelson, C. A., Butte, A. J. & Baranzini, S. E. Integrating biomedical research and electronic health records to create knowledge-based biologically meaningful machine-readable embeddings. Nat. Commun. 10, 3045 (2019).
Brunk, E. et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 36, 272–281 (2018).
Shen, X. et al. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-022-00999-8 (2023).
Li, X.-J. et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci. Transl Med. 5, 207ra142 (2013).
Zimmer, A. et al. The geometry of clinical labs and wellness states from deeply phenotyped humans. Nat. Commun. 12, 3578 (2021).
Sowjanya, A. M. & Mrudula, O. Effective treatment of imbalanced datasets in health care using modified SMOTE coupled with stacked deep learning algorithms. Appl. Nanosci. 13, 1829–1840 (2023).
Beckmann, N. D. et al. Multiscale causal networks identify VGF as a key regulator of Alzheimer’s disease. Nat. Commun. 11, 3942 (2020).
US Preventive Services Task Force. et al. Screening for cervical cancer: US Preventive Services Task Force recommendation statement. JAMA 320, 674–686 (2018).
Chou, R., Dana, T., Blazina, I., Daeges, M. & Jeanne, T. L. Statins for prevention of cardiovascular disease in adults: evidence report and systematic review for the US Preventive Services Task Force. JAMA 316, 2008–2024 (2016).
Tripp, S. & Grueber, M. Economic impact of the Human Genome Project (Batelle Memorial Institute, 2011).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Shilo, S. et al. 10K: a large-scale prospective longitudinal study in Israel. Eur. J. Epidemiol. 36, 1187–1194 (2021).
Smith, L. M. et al. Fluorescence detection in automated DNA sequence analysis. Nature 321, 674–679 (1986).
Church, G. M. & Kieffer-Higgins, S. Multiplex DNA sequencing. Science 240, 185–188 (1988).
Fodor, S. P. et al. Light-directed, spatially addressable parallel chemical synthesis. Science 251, 767–773 (1991).
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996).
Morris, K. N., Jensen, K. B., Julin, C. M., Weil, M. & Gold, L. High affinity ligands from in vitro selection: complex targets. Proc. Natl Acad. Sci. USA 95, 2902–2907 (1998).
Gullberg, M. et al. Cytokine detection by antibody-based proximity ligation. Proc. Natl Acad. Sci. USA 101, 8420–8424 (2004).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Ozaki, K. et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 32, 650–654 (2002).
Smith, C. A. et al. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 (2005).
Wishart, D. S. et al. HMDB: the human metabolome database. Nucleic Acids Res. 35, D521–D526 (2007).
1000 Genomes Project Consortium. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012).
Smarr, L. Quantifying your body: a how-to guide from a systems biology perspective. Biotechnol. J. 7, 980–991 (2012).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Cirulli, E. T. et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 29, 488–500.e2 (2019).
Hou, Y.-C. C. et al. Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proc. Natl Acad. Sci. USA 117, 3053–3062 (2020).
Mason, A. E. et al. Detection of COVID-19 using multimodal data from a wearable device: results from the first TemPredict Study. Sci. Rep. 12, 3463 (2022).
Benn, M., Watts, G. F., Tybjærg-Hansen, A. & Nordestgaard, B. G. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 37, 1384–1394 (2016).
Zubair, N. et al. Genetic predisposition impacts clinical changes in a lifestyle coaching program. Sci. Rep. 9, 6805 (2019).
Koletzko, B. et al. FADS1 and FADS2 polymorphisms modulate fatty acid metabolism and dietary impact on health. Annu. Rev. Nutr. 39, 21–44 (2019).
Ferguson, J. F. et al. NOS3 gene polymorphisms are associated with risk markers of cardiovascular disease, and interact with omega-3 polyunsaturated fatty acids. Atherosclerosis 211, 539–544 (2010).
Barton, J. C., Edwards, C. Q. & Acton, R. T. HFE gene: structure, function, mutations, and associated iron abnormalities. Gene 574, 179–192 (2015).
Feder, J. N. et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat. Genet. 13, 399–408 (1996).
Ahn, J. et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 19, 2739–2745 (2010).
Heianza, Y., Ma, W., Manson, J. E., Rexrode, K. M. & Qi, L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: a systematic review and meta-analysis of prospective studies. J. Am. Heart Assoc. 6, e004947 (2017).
El Rouby, N., Lima, J. J. & Johnson, J. A. Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin. Drug Metab. Toxicol. 14, 447–460 (2018).
Acknowledgements
The authors would like to thank N. Levine for the assistance with drafting figures, B. Barry and M. Simmons for the thorough editing, and L. Pflieger and B. Yurkovich for the useful discussions regarding scientific content.
Author information
Authors and Affiliations
Contributions
J.T.Y., S.J.E., N.R. and J.L.B. researched the literature. J.T.Y., S.J.E., N.R., J.L.B., J.C.L. & L.E.H. contributed substantially to the discussions of the content. J.T.Y., S.J.E., N.R., J.L.B., J.C.L. & L.E.H. wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
Phenome Health is a nonprofit research organization. The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Melissa A. Haendel, Jason L. Vassy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Data Commons: https://www.datacommons.org/
Nutrition for Precision Health: https://commonfund.nih.gov/nutritionforprecisionhealth
Glossary
- Actionable recommendations
-
Behavioural or clinical intervention strategies driven by data that can be pursued to reach specific health outcome.
- Aptamers
-
Short, single-stranded nucleic acid fragments (DNA or RNA) that selectively bind to specific target molecules such as proteins with variable affinities and off-target binding.
- Deep learning
-
A specific machine learning approach inspired by the human brain that uses multi-layered (so-called deep) network architectures.
- Deep phenotyping
-
A comprehensive analysis of phenotypic drivers and endpoints, from molecular assays and clinical data to social determinants of health and digital measures of environmental exposures and behaviours.
- Digital biomarkers
-
Lifestyle and physiological factors measured via digital health devices that enable remote — and possibly non-invasive — surveillance of the health state.
- Digital health
-
Digital tools such as mobile apps, wearables and telehealth that help provide a more holistic view of the health state of an individual.
- Digital twins
-
Computable digital, dynamic representations of a human that continuously integrates physiological and biochemical data with mechanistic knowledge.
- Explainable artificial intelligence
-
(XAI). Interpretable deep learning that provides explanations for black-box predictions.
- Genetic variants
-
Differences in the DNA sequence — from single nucleotides to large regions — among individuals, populations or species that may be inherited (passed down from parents in the germline) or somatic (developed de novo) and can explain differences in physical appearance, disease susceptibility and how people react to pharmacological interventions.
- Health outcomes
-
Labels derived from clinical metrics that reflect an individual’s dynamic state of physical, mental and social well-being, often used as ground truth (or expected values) to train computational models.
- Health trajectory
-
The trend of the integrated health of an individual, which is multidimensional and may simultaneously direct away from one disease and towards another.
- Incidental findings
-
Observations that were not the primary objective of the study but could have potential significance or implications for the health of the subjects.
- Knowledge graphs
-
Network representations of entities (for example, molecules, diseases and drugs) and the relationships between them.
- Multi-omics
-
Multiple omic data modalities, such as genomics, transcriptomics, epigenomics, metabolomics, microbiomics and proteomics.
- Personalized population health
-
An approach aimed at optimizing the health trajectory of each individual at the population scale.
- Phenome
-
The collection of dynamic and observable characteristics of an organism, ranging from the amount of markers in blood (such as cholesterol levels) to physiological signals (such as heart rate).
- Rare diseases
-
One of approximately 7,000 diseases affecting fewer than 1 in 200,000 people (USA) or fewer than 1 in 2,000 people (Europe), of which about 80% are thought to be genetic (so-called Mendelian diseases).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yurkovich, J.T., Evans, S.J., Rappaport, N. et al. The transition from genomics to phenomics in personalized population health. Nat Rev Genet 25, 286–302 (2024). https://doi.org/10.1038/s41576-023-00674-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-023-00674-x
This article is cited by
-
Global Healthspan Summit 2023: closing the gap between healthspan and lifespan
Nature Aging (2024)