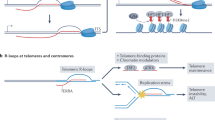
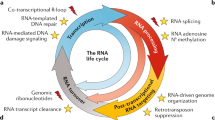
Abstract
Repetitive elements in the human genome, once considered ‘junk DNA’, are now known to adopt more than a dozen alternative (that is, non-B) DNA structures, such as self-annealed hairpins, left-handed Z-DNA, three-stranded triplexes (H-DNA) or four-stranded guanine quadruplex structures (G4 DNA). These dynamic conformations can act as functional genomic elements involved in DNA replication and transcription, chromatin organization and genome stability. In addition, recent studies have revealed a role for these alternative structures in triggering error-generating DNA repair processes, thereby actively enabling genome plasticity. As a driving force for genetic variation, non-B DNA structures thus contribute to both disease aetiology and evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022). The most recent and compete human genome sequencing assembly reveals more repetitive elements in the human genome than researchers have previously estimated, which could potentially support non-B DNA formation.
Plohl, M., Luchetti, A., Mestrovic, N. & Mantovani, B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409, 72–82 (2008).
Thakur, J., Packiaraj, J. & Henikoff, S. Sequence, chromatin and evolution of satellite DNA. Int. J. Mol. Sci. 22, 4309 (2021).
Herbert, A. ALU non-B-DNA conformations, flipons, binary codes and evolution. R. Soc. Open. Sci. 7, 200222 (2020).
Kasinathan, S. & Henikoff, S. Non-B-form DNA is enriched at centromeres. Mol. Biol. Evol. 35, 949–962 (2018).
Wang, G. & Vasquez, K. M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 19, 143–151 (2014).
Choi, J. & Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 40, 5893–5909 (2011).
Guiblet, W. M. et al. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 28, 1767–1778 (2018).
Marshall, P. R. et al. Dynamic regulation of Z-DNA in the mouse prefrontal cortex by the RNA-editing enzyme Adar1 is required for fear extinction. Nat. Neurosci. 23, 718–729 (2020). The ADAR1 protein binds to Z-DNA in the mouse prefrontal cortex during fear extinction learning and supresses or reduces Z-DNA formation, which is suggested to be required for memory flexibility.
Mirkin, S. M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
Praseuth, D., Guieysse, A. L. & Helene, C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta 1489, 181–206 (1999).
Huppert, J. L. & Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 (2007).
Marsico, G. et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874 (2019).
Valton, A. L. & Prioleau, M. N. G-quadruplexes in DNA replication: a problem or a necessity? Trends Genet. 32, 697–706 (2016).
Wang, G. & Vasquez, K. M. Effects of replication and transcription on DNA structure-related genetic instability. Genes 8, 17 (2017).
Prioleau, M. N. G-quadruplexes and DNA replication origins. Adv. Exp. Med. Biol. 1042, 273–286 (2017).
St Germain, C., Zhao, H. & Barlow, J. H. Transcription-replication collisions — a series of unfortunate events. Biomolecules 11, 1249 (2021).
Liu, G., Chen, X., Bissler, J. J., Sinden, R. R. & Leffak, M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 6, 652–659 (2010).
Gomes-Pereira, M., Fortune, M. T. & Monckton, D. G. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 10, 845–854 (2001).
Fu, Y. H. et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67, 1047–1058 (1991).
Kremer, E. J. et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 252, 1711–1714 (1991).
La Spada, A. R., Wilson, E. M., Lubahn, D. B., Harding, A. E. & Fischbeck, K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 (1991).
Catasus, L. et al. Frameshift mutations at coding mononucleotide repeat microsatellites in endometrial carcinoma with microsatellite instability. Cancer 88, 2290–2297 (2000).
Georgakopoulos-Soares, I. et al. Transcription-coupled repair and mismatch repair contribute towards preserving genome integrity at mononucleotide repeat tracts. Nat. Commun. 11, 1980 (2020). Using bioinformatic approaches, this study reports transcription-associated asymmetrical distribution of repetitive elements, insertions and deletions at repeats in human cancer genomes, with involvement of DNA repair pathways.
Rothenburg, S., Koch-Nolte, F., Rich, A. & Haag, F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA 98, 8985–8990 (2001).
Malik, I., Kelley, C. P., Wang, E. T. & Todd, P. K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607 (2021).
Paulson, H. L. & Fischbeck, K. H. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 19, 79–107 (1996).
McMurray, C. T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11, 786–799 (2010).
Jones, L., Houlden, H. & Tabrizi, S. J. DNA repair in the trinucleotide repeat disorders. Lancet Neurol. 16, 88–96 (2017).
Cleary, J. D. & Pearson, C. E. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 21, 272–280 (2005).
McKinney, J. A. et al. Distinct DNA repair pathways cause genomic instability at alternative DNA structures. Nat. Commun. 11, 236 (2020). This study reports that the MMR protein complex MSH2–MSH3 binds to Z-DNA and recruits the NER nuclease ERCC1–XPF to the site, resulting in structure-specific cleavage and DSBs at Z-DNA regardless of DNA replication status.
Zhao, J. et al. Distinct mechanisms of nuclease-directed DNA-structure-induced genetic instability in cancer genomes. Cell Rep. 22, 1200–1210 (2018).
Iyer, R. R., Pluciennik, A., Napierala, M. & Wells, R. D. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 84, 199–226 (2015).
Sundararajan, R. & Freudenreich, C. H. Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in Saccharomyces cerevisiae. PLoS Genet. 7, e1001339 (2011). Long CAG/CTG repeats trigger an MRX-dependent DNA damage checkpoint response in budding yeast, which affects the cell cycle, leading to repeat-dependent S-phase delays and G2/M arrests, which results in morphological abnormalities.
Voineagu, I., Surka, C. F., Shishkin, A. A., Krasilnikova, M. M. & Mirkin, S. M. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat. Struct. Mol. Biol. 16, 226–228 (2009).
Orr, H. T. & Zoghbi, H. Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621 (2007).
Ye, C., Ji, G., Li, L. & Liang, C. detectIR: a novel program for detecting perfect and imperfect inverted repeats using complex numbers and vector calculation. PLoS ONE 9, e113349 (2014).
Kikin, O., D’Antonio, L. & Bagga, P. S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–W682 (2006).
Brazda, V. et al. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics 35, 3493–3495 (2019).
Wang, G., Gaddis, S. & Vasquez, K. M. Methods to detect replication-dependent and replication-independent DNA structure-induced genetic instability. Methods 64, 67–72 (2013).
Barshai, M., Aubert, A. & Orenstein, Y. G4detector: convolutional neural network to predict DNA G-quadruplexes. IEEE/ACM Trans. Comput. Biol. Bioinform. 19, 1946–1955 (2022).
Jenjaroenpun, P. & Kuznetsov, V. A. TTS mapping: integrative WEB tool for analysis of triplex formation target DNA sequences, G-quadruplets and non-protein coding regulatory DNA elements in the human genome. BMC Genomics 10, S9 (2009).
Cer, R. Z. et al. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 41, D94–D100 (2013).
Wang, G., Zhao, J. & Vasquez, K. M. Detection of cis- and trans-acting factors in DNA structure-induced genetic instability using in silico and cellular approaches. Front. Genet. 7, 135 (2016).
Beknazarov, N., Jin, S. & Poptsova, M. Deep learning approach for predicting functional Z-DNA regions using omics data. Sci. Rep. 10, 19134 (2020).
Rocher, V., Genais, M., Nassereddine, E. & Mourad, R. DeepG4: a deep learning approach to predict cell-type specific active G-quadruplex regions. PLoS Comput. Biol. 17, e1009308 (2021).
Bian, Y. et al. Insights into the kinetic partitioning folding dynamics of the human telomeric G-quadruplex from molecular simulations and machine learning. J. Chem. Theory Comput. 16, 5936–5947 (2020).
Kouzine, F. et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 4, 344–356 e347 (2017).
Abeysinghe, S. S., Chuzhanova, N., Krawczak, M., Ball, E. V. & Cooper, D. N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 22, 229–244 (2003).
Rahmouni, A. R. & Wells, R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science 246, 358–363 (1989).
Koeris, M., Funke, L., Shrestha, J., Rich, A. & Maas, S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 33, 5362–5370 (2005).
Herbert, A. et al. The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 26, 3486–3493 (1998).
Zhang, X., Spiegel, J., Martinez Cuesta, S., Adhikari, S. & Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 13, 626–633 (2021). Hundreds of putative G4 DNA binding proteins from various functional classes are identified using G4-ligand probes crosslinked to G4 binding proteins in situ in living cells, suggesting complex and active DNA structure-related metabolism in vivo.
Zheng, L. L. et al. pH-responsive DNA motif: from rational design to analytical applications. Front. Chem. 9, 732770 (2021).
Son, H., Bae, S. & Lee, S. A thermodynamic understanding of the salt-induced B-to-Z transition of DNA containing BZ junctions. Biochem. Biophys. Res. Commun. 583, 142–145 (2021).
Potaman, V. N., Ussery, D. W. & Sinden, R. R. Formation of a combined H-DNA/open TATA box structure in the promoter sequence of the human Na,K-ATPase alpha2 gene. J. Biol. Chem. 271, 13441–13447 (1996).
Htun, H. & Dahlberg, J. E. Topology and formation of triple-stranded H-DNA. Science 243, 1571–1576 (1989).
Latha, K. S., Anitha, S., Rao, K. S. & Viswamitra, M. A. Molecular understanding of aluminum-induced topological changes in (CCG)12 triplet repeats: relevance to neurological disorders. Biochim. Biophys. Acta 1588, 56–64 (2002).
Fakharzadeh, A., Zhang, J., Roland, C. & Sagui, C. Novel eGZ-motif formed by regularly extruded guanine bases in a left-handed Z-DNA helix as a major motif behind CGG trinucleotide repeats. Nucleic Acids Res. 50, 4860–4876 (2022).
Ajjugal, Y., Kolimi, N. & Rathinavelan, T. Secondary structural choice of DNA and RNA associated with CGG/CCG trinucleotide repeat expansion rationalizes the RNA misprocessing in FXTAS. Sci. Rep. 11, 8163 (2021).
Kim, S. H., Jung, H. J., Lee, I. B., Lee, N. K. & Hong, S. C. Sequence-dependent cost for Z-form shapes the torsion-driven B-Z transition via close interplay of Z-DNA and DNA bubble. Nucleic Acids Res. 49, 3651–3660 (2021).
Zhang, F., Huang, Q., Yan, J. & Chen, Z. Histone acetylation induced transformation of B-DNA to Z-DNA in cells probed through FT-IR spectroscopy. Anal. Chem. 88, 4179–4182 (2016).
Li, Y. et al. Remodeling chromatin induces Z-DNA conformation detected through Fourier transform infrared spectroscopy. Anal. Chem. 92, 14452–14458 (2020).
Krassovsky, K., Ghosh, R. P. & Meyer, B. J. Genome-wide profiling reveals functional interplay of DNA sequence composition, transcriptional activity, and nucleosome positioning in driving DNA supercoiling and helix destabilization in C. elegans. Genome Res. 31, 1187–1202 (2021). DNA supercoiling regions and non-B DNA structures in the genome of C. elegans embryos are mapped and found to co-localize at functional regions in the genome, such as transcription start sites.
Wittig, B., Dorbic, T. & Rich, A. Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei. Proc. Natl Acad. Sci. USA 88, 2259–2263 (1991).
Wolfl, S., Wittig, B. & Rich, A. Identification of transcriptionally induced Z-DNA segments in the human c-myc gene. Biochim. Biophys. Acta 1264, 294–302 (1995).
Wittig, B., Wolfl, S., Dorbic, T., Vahrson, W. & Rich, A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 11, 4653–4663 (1992).
Michelotti, G. A. et al. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16, 2656–2669 (1996).
Feng, X., Xie, F. Y., Ou, X. H. & Ma, J. Y. Cruciform DNA in mouse growing oocytes: its dynamics and its relationship with DNA transcription. PLoS ONE 15, e0240844 (2020).
Schwartz, T., Behlke, J., Lowenhaupt, K., Heinemann, U. & Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 (2001).
Baik, J. Y. et al. ZBP1 not RIPK1 mediates tumor necroptosis in breast cancer. Nat. Commun. 12, 2666 (2021).
Ha, S. C. et al. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc. Natl Acad. Sci. USA 105, 20671–20676 (2008).
Rothenburg, S. et al. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl Acad. Sci. USA 102, 1602–1607 (2005).
Kim, Y. G., Lowenhaupt, K., Oh, D. B., Kim, K. K. & Rich, A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: implications for development of a therapy for poxvirus infection. Proc. Natl Acad. Sci. USA 101, 1514–1518 (2004).
Kus, K. et al. The structure of the Cyprinid herpesvirus 3 ORF112-Zalpha.Z-DNA complex reveals a mechanism of nucleic acids recognition conserved with E3L, a poxvirus inhibitor of interferon response. J. Biol. Chem. 290, 30713–30725 (2015).
Brazda, V., Laister, R. C., Jagelska, E. B. & Arrowsmith, C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 12, 33 (2011).
Reddy, M. C., Christensen, J. & Vasquez, K. M. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry 44, 4188–4195 (2005).
Meier-Stephenson, V. G4-quadruplex-binding proteins: review and insights into selectivity. Biophys. Rev. 14, 635–654 (2022).
Chu, W. K. & Hickson, I. D. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9, 644–654 (2009).
Liberi, G. et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 (2005).
Shishkin, A. A. et al. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol. Cell 35, 82–92 (2009).
Butler, D. K., Yasuda, L. E. & Yao, M. C. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell 87, 1115–1122 (1996).
Fleming, A. M., Ding, Y. & Burrows, C. J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl Acad. Sci. USA 114, 2604–2609 (2017).
Fleming, A. M., Zhu, J., Jara-Espejo, M. & Burrows, C. J. Cruciform DNA sequences in gene promoters can impact transcription upon oxidative modification of 2′-deoxyguanosine. Biochemistry 59, 2616–2626 (2020).
Roychoudhury, S. et al. Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome. Proc. Natl Acad. Sci. USA 117, 11409–11420 (2020).
Volker, J., Plum, G. E., Klump, H. H. & Breslauer, K. J. DNA repair and DNA triplet repeat expansion: the impact of abasic lesions on triplet repeat DNA energetics. J. Am. Chem. Soc. 131, 9354–9360 (2009).
Lai, Y. et al. Base excision repair of chemotherapeutically-induced alkylated DNA damage predominantly causes contractions of expanded GAA repeats associated with Friedreich’s ataxia. PLoS ONE 9, e93464 (2014).
Bochman, M. L., Paeschke, K. & Zakian, V. A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 (2012).
Smith, S. S. Evolutionary expansion of structurally complex DNA sequences. Cancer Genomics Proteomics 7, 207–215 (2010).
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Godde, J. S., Kass, S. U., Hirst, M. C. & Wolffe, A. P. Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J. Biol. Chem. 271, 24325–24328 (1996).
Linxweller, W. & Horz, W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell 42, 281–290 (1985).
Ruan, H. & Wang, Y. H. Friedreich’s ataxia GAA.TTC duplex and GAA.GAA.TTC triplex structures exclude nucleosome assembly. J. Mol. Biol. 383, 292–300 (2008).
Miura, O., Ogake, T., Yoneyama, H., Kikuchi, Y. & Ohyama, T. A strong structural correlation between short inverted repeat sequences and the polyadenylation signal in yeast and nucleosome exclusion by these inverted repeats. Curr. Genet. 65, 575–590 (2019).
Wong, H. M. & Huppert, J. L. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 5, 1713–1719 (2009).
Shen, J. et al. Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol. 22, 143 (2021).
Godde, J. S. & Wolffe, A. P. Nucleosome assembly on CTG triplet repeats. J. Biol. Chem. 271, 15222–15229 (1996).
Wada-Kiyama, Y. & Kiyama, R. Conservation and periodicity of DNA bend sites in the human beta-globin gene locus. J. Biol. Chem. 270, 12439–12445 (1995).
Hou, Y. et al. Integrative characterization of G-quadruplexes in the three-dimensional chromatin structure. Epigenetics 14, 894–911 (2019).
Kanoh, Y. et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 22, 889–897 (2015). Rif1 is able to bind to G4 DNA motifs at selected intergenic regions in the fission yeast genome and create local chromatin structures that suppress late-firing of dormant origins located up to 50 kb from these regions.
Barbault, F., Huynh-Dinh, T., Paoletti, J. & Lanceloti, G. A new peculiar DNA structure: NMR solution structure of a DNA kissing complex. J. Biomol. Struct. Dyn. 19, 649–658 (2002).
Xu, X. & Chen, S. J. Topological constraints of RNA pseudoknotted and loop-kissing motifs: applications to three-dimensional structure prediction. Nucleic Acids Res. 48, 6503–6512 (2020).
Williams, J. D. et al. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop ‘G4 Kissing’ interaction. Nucleic Acids Res. 48, 5907–5925 (2020).
Son, L. S., Bacolla, A. & Wells, R. D. Sticky DNA: in vivo formation in E. coli and in vitro association of long GAA*TTC tracts to generate two independent supercoiled domains. J. Mol. Biol. 360, 267–284 (2006).
Vetcher, A. A., Napierala, M. & Wells, R. D. Sticky DNA: effect of the polypurine.polypyrimidine sequence. J. Biol. Chem. 277, 39228–39234 (2002).
Vanaja, A. & Yella, V. R. Delineation of the DNA structural features of eukaryotic core promoter classes. ACS Omega 7, 5657–5669 (2022).
Hershman, S. G. et al. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 36, 144–156 (2008).
Miura, O., Ogake, T. & Ohyama, T. Requirement or exclusion of inverted repeat sequences with cruciform-forming potential in Escherichia coli revealed by genome-wide analyses. Curr. Genet. 64, 945–958 (2018).
Du, X. et al. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 41, 5965–5977 (2013).
Drew, H. R., Weeks, J. R. & Travers, A. A. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 4, 1025–1032 (1985).
Wang, J. C. & Lynch, A. S. Transcription and DNA supercoiling. Curr. Opin. Genet. Dev. 3, 764–768 (1993).
Mizutani, M., Ohta, T., Watanabe, H., Handa, H. & Hirose, S. Negative supercoiling of DNA facilitates an interaction between transcription factor IID and the fibroin gene promoter. Proc. Natl Acad. Sci. USA 88, 718–722 (1991).
Aboul-ela, F., Bowater, R. P. & Lilley, D. M. Competing B-Z and helix-coil conformational transitions in supercoiled plasmid DNA. J. Biol. Chem. 267, 1776–1785 (1992).
Varshney, D., Spiegel, J., Zyner, K., Tannahill, D. & Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 21, 459–474 (2020).
Lago, S. et al. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 12, 3885 (2021).
Revikumar, A. et al. Multiple G-quadruplex binding ligand induced transcriptomic map of cancer cell lines. J. Cell Commun. Signal. 16, 129–135 (2022).
Ditlevson, J. V. et al. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 36, 3163–3170 (2008).
Belotserkovskii, B. P. et al. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 282, 32433–32441 (2007).
Pandey, S. et al. Transcription blockage by stable H-DNA analogs in vitro. Nucleic Acids Res. 43, 6994–7004 (2015).
Xu, J., Chong, J. & Wang, D. Opposite roles of transcription elongation factors Spt4/5 and Elf1 in RNA polymerase II transcription through B-form versus non-B DNA structures. Nucleic Acids Res. 49, 4944–4953 (2021).
Belotserkovskii, B. P. et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl Acad. Sci. USA 107, 12816–12821 (2010).
Agarwal, T., Roy, S., Kumar, S., Chakraborty, T. K. & Maiti, S. In the sense of transcription regulation by G-quadruplexes: asymmetric effects in sense and antisense strands. Biochemistry 53, 3711–3718 (2014).
Tsai, Z. T., Chu, W. Y., Cheng, J. H. & Tsai, H. K. Associations between intronic non-B DNA structures and exon skipping. Nucleic Acids Res. 42, 739–747 (2014).
Darnell, J. E. Jr. Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science 202, 1257–1260 (1978).
Nieto Moreno, N., Giono, L. E., Cambindo Botto, A. E., Munoz, M. J. & Kornblihtt, A. R. Chromatin, DNA structure and alternative splicing. FEBS Lett. 589, 3370–3378 (2015).
Dai, X. & Rothman-Denes, L. B. DNA structure and transcription. Curr. Opin. Microbiol. 2, 126–130 (1999).
Kim, N. The Interplay between G-quadruplex and transcription. Curr. Med. Chem. 26, 2898–2917 (2019).
Samadashwily, G. M., Raca, G. & Mirkin, S. M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17, 298–304 (1997).
Kim, Y. S. & Kang, H. S. Sequence-specific functions of the early palindrome domain within the SV40 core origin of replication. Nucleic Acids Res. 17, 9279–9289 (1989).
Lin, S. & Kowalski, D. DNA helical instability facilitates initiation at the SV40 replication origin. J. Mol. Biol. 235, 496–507 (1994).
Pearson, C. E., Zorbas, H., Price, G. B. & Zannis-Hadjopoulos, M. Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J. Cell. Biochem. 63, 1–22 (1996).
Lerner, L. K. & Sale, J. E. Replication of G quadruplex DNA. Genes 10, 95 (2019).
Guilbaud, G. et al. Determination of human DNA replication origin position and efficiency reveals principles of initiation zone organisation. Nucleic Acids Res. 50, 7436–7450 (2022).
Bartholdy, B., Mukhopadhyay, R., Lajugie, J., Aladjem, M. I. & Bouhassira, E. E. Allele-specific analysis of DNA replication origins in mammalian cells. Nat. Commun. 6, 7051 (2015).
Schneider, T. D. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 29, 4881–4891 (2001).
Valton, A. L. et al. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 33, 732–746 (2014).
Yahyaoui, W., Callejo, M., Price, G. B. & Zannis-Hadjopoulos, M. Deletion of the cruciform binding domain in CBP/14-3-3 displays reduced origin binding and initiation of DNA replication in budding yeast. BMC Mol. Biol. 8, 27 (2007).
Hoshina, S. et al. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J. Biol. Chem. 288, 30161–30171 (2013).
Prorok, P. et al. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun. 10, 3274 (2019). G4 DNA-forming sequences in the OGRE are required for the activity of several types of replication origin; adding G4 DNA-stabilizing ligands affects origin activities accordingly, suggesting a role for G4 DNA in replication regulation.
Hile, S. E. & Eckert, K. A. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J. Mol. Biol. 335, 745–759 (2004).
Anand, R. P. et al. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 40, 1091–1105 (2012).
Wang, Q. et al. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 39, 6229–6237 (2011).
Paeschke, K., Capra, J. A. & Zakian, V. A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 (2011).
Kopel, V., Pozner, A., Baran, N. & Manor, H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 24, 330–335 (1996).
Yangyuoru, P. M., Bradburn, D. A., Liu, Z., Xiao, T. S. & Russell, R. The G-quadruplex (G4) resolvase DHX36 efficiently and specifically disrupts DNA G4s via a translocation-based helicase mechanism. J. Biol. Chem. 293, 1924–1932 (2018).
Le, T. T. et al. Synergistic coordination of chromatin torsional mechanics and topoisomerase activity. Cell 179, 619–631.e15 (2019).
Yuan, Z. et al. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 11, 688 (2020).
Masuda-Sasa, T., Polaczek, P., Peng, X. P., Chen, L. & Campbell, J. L. Processing of G4 DNA by DNA2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of DNA2/RPA substrate recognition. J. Biol. Chem. 283, 24359–24373 (2008).
Peleg, M., Kopel, V., Borowiec, J. A. & Manor, H. Formation of DNA triple helices inhibits DNA unwinding by the SV40 large T-antigen helicase. Nucleic Acids Res. 23, 1292–1299 (1995).
Lopes, J. et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 30, 4033–4046 (2011).
Dovrat, D. et al. A live-cell imaging approach for measuring DNA replication rates. Cell Rep. 24, 252–258 (2018).
Kobori, J. A., Strauss, E., Minard, K. & Hood, L. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science 234, 173–179 (1986).
Weinreb, A., Collier, D. A., Birshtein, B. K. & Wells, R. D. Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J. Biol. Chem. 265, 1352–1359 (1990).
Vallur, A. C. & Maizels, N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc. Natl Acad. Sci. USA 105, 16508–16512 (2008).
Wahls, W. P., Wallace, L. J. & Moore, P. D. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol. 10, 785–793 (1990).
Roy, U. & Greene, E. C. Demystifying the D-loop during DNA recombination. Nature 586, 677–678 (2020).
Haniford, D. B. & Pulleyblank, D. E. The in-vivo occurrence of Z DNA. J. Biomol. Struct. Dyn. 1, 593–609 (1983).
Blaho, J. A. & Wells, R. D. Left-handed Z-DNA and genetic recombination. Prog. Nucleic Acid Res. Mol. Biol. 37, 107–126 (1989).
Xu, Z., Zan, H., Pone, E. J., Mai, T. & Casali, P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 12, 517–531 (2012).
Larson, E. D., Duquette, M. L., Cummings, W. J., Streiff, R. J. & Maizels, N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 15, 470–474 (2005).
Tashiro, J., Kinoshita, K. & Honjo, T. Palindromic but not G-rich sequences are targets of class switch recombination. Int. Immunol. 13, 495–505 (2001).
Guiblet, W. M. et al. Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome. Nucleic Acids Res. 49, 1497–1516 (2021).
Georgakopoulos-Soares, I., Morganella, S., Jain, N., Hemberg, M. & Nik-Zainal, S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 28, 1264–1271 (2018).
Thornton, C. A., Johnson, K. & Moxley, R. T. III Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 35, 104–107 (1994).
Zatz, M. et al. Analysis of the CTG repeat in skeletal muscle of young and adult myotonic dystrophy patients: when does the expansion occur? Hum. Mol. Genet. 4, 401–406 (1995).
Rider, S. D.Jr et al. Stable G-quadruplex DNA structures promote replication-dependent genome instability. J. Biol. Chem. 298, 101947 (2022).
Lu, S. et al. Short inverted repeats are hotspots for genetic instability: relevance to cancer genomes. Cell Rep. 10, 1674–1680 (2015).
Wang, G., Christensen, L. A. & Vasquez, K. M. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl Acad. Sci. USA 103, 2677–2682 (2006).
Wang, G. & Vasquez, K. M. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl Acad. Sci. USA 101, 13448–13453 (2004).
Takahashi, S., Brazier, J. A. & Sugimoto, N. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Natl Acad. Sci. USA 114, 9605–9610 (2017).
Burrow, A. A., Marullo, A., Holder, L. R. & Wang, Y. H. Secondary structure formation and DNA instability at fragile site FRA16B. Nucleic Acids Res. 38, 2865–2877 (2010).
Murat, P., Guilbaud, G. & Sale, J. E. DNA polymerase stalling at structured DNA constrains the expansion of short tandem repeats. Genome Biol. 21, 209 (2020).
Kamath-Loeb, A. S., Loeb, L. A., Johansson, E., Burgers, P. M. & Fry, M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 276, 16439–16446 (2001).
Shah, S. N., Opresko, P. L., Meng, X., Lee, M. Y. & Eckert, K. A. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 38, 1149–1162 (2010).
Lormand, J. D. et al. DNA polymerase delta stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 41, 10323–10333 (2013).
Iyer, R. R., Pluciennik, A., Rosche, W. A., Sinden, R. R. & Wells, R. D. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J. Biol. Chem. 275, 2174–2184 (2000).
Teng, F. Y. et al. Escherichia coli DNA polymerase I can disrupt G-quadruplex structures during DNA replication. FEBS J. 284, 4051–4065 (2017).
Shah, K. A. et al. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2, 1088–1095 (2012).
Abdulovic, A. L., Hile, S. E., Kunkel, T. A. & Eckert, K. A. The in vitro fidelity of yeast DNA polymerase delta and polymerase epsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA Repair 10, 497–505 (2011).
Guo, J., Gu, L., Leffak, M. & Li, G. M. MutSbeta promotes trinucleotide repeat expansion by recruiting DNA polymerase beta to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 26, 775–786 (2016).
Meyer, D. et al. Cooperation between non-essential DNA polymerases contributes to genome stability in Saccharomyces cerevisiae. DNA Repair 76, 40–49 (2019).
Eddy, S. et al. Human translesion polymerase kappa exhibits enhanced activity and reduced fidelity two nucleotides from G-quadruplex DNA. Biochemistry 55, 5218–5229 (2016).
Betous, R. et al. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 48, 369–378 (2009).
Eddy, S. et al. Evidence for the kinetic partitioning of polymerase activity on G-quadruplex DNA. Biochemistry 54, 3218–3230 (2015).
Gadgil, R. Y. et al. Replication stress at microsatellites causes DNA double-strand breaks and break-induced replication. J. Biol. Chem. 295, 15378–15397 (2020).
Stern, H. R., Sefcikova, J., Chaparro, V. E. & Beuning, P. J. Mammalian DNA polymerase kappa activity and specificity. Molecules 24, 2805 (2019).
Walsh, E., Wang, X., Lee, M. Y. & Eckert, K. A. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J. Mol. Biol. 425, 232–243 (2013).
Twayana, S. et al. Translesion polymerase eta both facilitates DNA replication and promotes increased human genetic variation at common fragile sites. Proc. Natl Acad. Sci. USA 118, e2106477118 (2021).
Ketkar, A. et al. Human Rev1 relies on insert-2 to promote selective binding and accurate replication of stabilized G-quadruplex motifs. Nucleic Acids Res. 49, 2065–2084 (2021).
Northam, M. R. et al. DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 42, 290–306 (2014).
Sarkies, P., Reams, C., Simpson, L. J. & Sale, J. E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713 (2010).
Koole, W. et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 5, 3216 (2014).
Chan, K. Y., Li, X., Ortega, J., Gu, L. & Li, G. M. DNA polymerase theta promotes CAG*CTG repeat expansions in Huntington’s disease via insertion sequences of its catalytic domain. J. Biol. Chem. 297, 101144 (2021).
Boyer, A. S., Grgurevic, S., Cazaux, C. & Hoffmann, J. S. The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. J. Mol. Biol. 425, 4767–4781 (2013).
Lemmens, B., van Schendel, R. & Tijsterman, M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 6, 8909 (2015).
Kang, S., Jaworski, A., Ohshima, K. & Wells, R. D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10, 213–218 (1995).
Cleary, J. D., Nichol, K., Wang, Y. H. & Pearson, C. E. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 31, 37–46 (2002).
Lia, A. S. et al. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet. 7, 1285–1291 (1998).
Telenius, H. et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 6, 409–414 (1994).
Hashida, H., Goto, J., Kurisaki, H., Mizusawa, H. & Kanazawa, I. Brain regional differences in the expansion of a CAG repeat in the spinocerebellar ataxias: dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and spinocerebellar ataxia type 1. Ann. Neurol. 41, 505–511 (1997).
Liu, T., Luo, H. & Gao, F. Position preference of essential genes in prokaryotic operons. PLoS ONE 16, e0250380 (2021).
Prioleau, M. N. & MacAlpine, D. M. DNA replication origins-where do we begin? Genes Dev. 30, 1683–1697 (2016).
Thys, R. G., Lehman, C. E., Pierce, L. C. & Wang, Y. H. DNA secondary structure at chromosomal fragile sites in human disease. Curr. Genomics 16, 60–70 (2015).
Arlt, M. F., Durkin, S. G., Ragland, R. L. & Glover, T. W. Common fragile sites as targets for chromosome rearrangements. DNA Repair 5, 1126–1135 (2006).
Sutherland, G. R. Rare fragile sites. Cytogenet. Genome Res. 100, 77–84 (2003).
Glover, T. W. Instability at chromosomal fragile sites. Recent Results Cancer Res. 154, 185–199 (1998).
Brison, O. et al. Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide. Nat. Commun. 10, 5693 (2019).
Smith, D. I., Zhu, Y., McAvoy, S. & Kuhn, R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 232, 48–57 (2006).
Helmrich, A., Ballarino, M. & Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011).
Sankar, T. S., Wastuwidyaningtyas, B. D., Dong, Y., Lewis, S. A. & Wang, J. D. The nature of mutations induced by replication–transcription collisions. Nature 535, 178–181 (2016). Replication–transcription collisions in the genome of actively dividing bacterial cells result in duplications and deletions at sites of replication stalling where replication forks enter a transcription unit, resulting in T>C base substitutions on the non-template strand, not only at the sites of collision but also in adjacent areas.
Kim, N. & Jinks-Robertson, S. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459, 1150–1153 (2009).
Macheret, M. & Halazonetis, T. D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10, 425–448 (2015).
Buschta-Hedayat, N., Buterin, T., Hess, M. T., Missura, M. & Naegeli, H. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc. Natl Acad. Sci. USA 96, 6090–6095 (1999).
Majchrzak, M., Bowater, R. P., Staczek, P. & Parniewski, P. SOS repair and DNA supercoiling influence the genetic stability of DNA triplet repeats in Escherichia coli. J. Mol. Biol. 364, 612–624 (2006).
Lahiri, M., Gustafson, T. L., Majors, E. R. & Freudenreich, C. H. Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol. Cell 15, 287–293 (2004).
Bacolla, A., Jaworski, A., Connors, T. D. & Wells, R. D. Pkd1 unusual DNA conformations are recognized by nucleotide excision repair. J. Biol. Chem. 276, 18597–18604 (2001).
Tran, H., Degtyareva, N., Gordenin, D. & Resnick, M. A. Altered replication and inverted repeats induce mismatch repair-independent recombination between highly diverged DNAs in yeast. Mol. Cell. Biol. 17, 1027–1036 (1997).
Nag, D. K. & Kurst, A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics 146, 835–847 (1997).
Bill, C. A., Taghian, D. G., Duran, W. A. & Nickoloff, J. A. Repair bias of large loop mismatches during recombination in mammalian cells depends on loop length and structure. Mutat. Res. 485, 255–265 (2001).
Panigrahi, G. B., Lau, R., Montgomery, S. E., Leonard, M. R. & Pearson, C. E. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 12, 654–662 (2005).
Palombo, F. et al. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6, 1181–1184 (1996).
Owen, B. A. et al. CAG)(n)-hairpin DNA binds to Msh2–Msh3 and changes properties of mismatch recognition. Nat. Struct. Biol. 12, 663–670 (2005).
Manley, K., Shirley, T. L., Flaherty, L. & Messer, A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23, 471–473 (1999).
van den Broek, W. J. et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11, 191–198 (2002).
Savouret, C. et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 22, 2264–2273 (2003).
Lin, Y., Dion, V. & Wilson, J. H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 13, 179–180 (2006).
Zhao, J., Jain, A., Iyer, R. R., Modrich, P. L. & Vasquez, K. M. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 37, 4420–4429 (2009).
Potaman, V. N. et al. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 32, 1224–1231 (2004).
Kim, H. M. et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 27, 2896–2906 (2008).
Neil, A. J. et al. Replication-independent instability of Friedreich’s ataxia GAA repeats during chronological aging. Proc. Natl Acad. Sci. USA 118, e2013080118 (2021). This study finds that MMR proteins are involved in generating DBSs at long GAA(100) repeats that can form H-DNA in non-dividing cells, resulting in large-scale deletions, including the repeat and adjacent regions, and mediated by error-prone non-homologous end-joining or gene conversions via ectopic homologous recombination.
Ehrat, E. A., Johnson, B. R., Williams, J. D., Borchert, G. M. & Larson, E. D. G-quadruplex recognition activities of E. coli MutS. BMC Mol. Biol. 13, 23 (2012).
Pavlova, A. V. et al. Responses of DNA mismatch repair proteins to a stable G-quadruplex embedded into a DNA duplex structure. Int. J. Mol. Sci. 21, 8773 (2020).
Lai, Y. et al. Crosstalk between MSH2-MSH3 and polbeta promotes trinucleotide repeat expansion during base excision repair. Nat. Commun. 7, 12465 (2016). The MMR protein complex MSH2–MSH3 is found to crosstalk with the BER machinery, stimulating the synthesis activity of DNA Polβ through triplet repeats and facilitating the formation of flap structures, which leads to repeat expansions.
McKinney, J. A., Wang, G. & Vasquez, K. M. Distinct mechanisms of mutagenic processing of alternative DNA structures by repair proteins. Mol. Cell Oncol. 7, 1743807 (2020).
Wood, R. D. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie 81, 39–44 (1999).
Wang, G., Seidman, M. M. & Glazer, P. M. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science 271, 802–805 (1996).
Vasquez, K. M., Christensen, J., Li, L., Finch, R. A. & Glazer, P. M. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad. Sci. USA 99, 5848–5853 (2002).
Thoma, B. S., Wakasugi, M., Christensen, J., Reddy, M. C. & Vasquez, K. M. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 33, 2993–3001 (2005).
Oussatcheva, E. A., Hashem, V. I., Zou, Y., Sinden, R. R. & Potaman, V. N. Involvement of the nucleotide excision repair protein UvrA in instability of CAG*CTG repeat sequences in Escherichia coli. J. Biol. Chem. 276, 30878–30884 (2001).
Szwarocka, S. T., Staczek, P. & Parniewski, P. Chromosomal model for analysis of a long CTG/CAG tract stability in wild-type Escherichia coli and its nucleotide excision repair mutants. Can. J. Microbiol. 53, 860–868 (2007).
Parniewski, P., Bacolla, A., Jaworski, A. & Wells, R. D. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 27, 616–623 (1999).
Matson, S. W. & Robertson, A. B. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 34, 4089–4097 (2006).
Lin, Y. & Wilson, J. H. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 27, 6209–6217 (2007).
Trujillo, K. M. & Sung, P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276, 35458–35464 (2001).
Farah, J. A., Hartsuiker, E., Mizuno, K., Ohta, K. & Smith, G. R. A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161, 461–468 (2002).
Paull, T. T., Cortez, D., Bowers, B., Elledge, S. J. & Gellert, M. Direct DNA binding by Brca1. Proc. Natl Acad. Sci. USA 98, 6086–6091 (2001).
De la Torre, C., Pincheira, J. & Lopez-Saez, J. F. Human syndromes with genomic instability and multiprotein machines that repair DNA double-strand breaks. Histol. Histopathol. 18, 225–243 (2003).
Lengsfeld, B. M., Rattray, A. J., Bhaskara, V., Ghirlando, R. & Paull, T. T. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651 (2007).
Ghosal, G. & Muniyappa, K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 33, 4692–4703 (2005).
Farah, J. A., Cromie, G., Steiner, W. W. & Smith, G. R. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 169, 1261–1274 (2005).
Jankowski, C. & Nag, D. K. Most meiotic CAG repeat tract-length alterations in yeast are SPO11 dependent. Mol. Genet. Genomics 267, 64–70 (2002).
Mankouri, H. W., Ashton, T. M. & Hickson, I. D. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc. Natl Acad. Sci. USA 108, 4944–4949 (2011).
Olmezer, G. et al. Replication intermediates that escape Dna2 activity are processed by Holliday junction resolvase Yen1. Nat. Commun. 7, 13157 (2016).
Wyatt, H. D., Sarbajna, S., Matos, J. & West, S. C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell 52, 234–247 (2013).
Xu, X. et al. Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex. Nucleic Acids Res. 49, 7740–7752 (2021).
Ashton, T. M., Mankouri, H. W., Heidenblut, A., McHugh, P. J. & Hickson, I. D. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 31, 1921–1933 (2011).
Agostinho, A. et al. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9, e1003591 (2013).
Cote, A. G. & Lewis, S. M. Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S. cerevisiae. Mol. Cell 31, 800–812 (2008).
Minocherhomji, S. & Hickson, I. D. Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol. 24, 321–327 (2014).
Ait Saada, A. et al. Structural parameters of palindromic repeats determine the specificity of nuclease attack of secondary structures. Nucleic Acids Res. 49, 3932–3947 (2021).
Goold, R. et al. FAN1 controls mismatch repair complex assembly via MLH1 retention to stabilize CAG repeat expansion in Huntington’s disease. Cell Rep. 36, 109649 (2021). The DNA-structure-specific nuclease FAN1 binds to the MMR protein MLH1 and suppresses its interaction with MSH3, thereby reducing MMR-promoted CAG repeat expansion in human cells.
Deshmukh, A. L. et al. FAN1 exo- not endo-nuclease pausing on disease-associated slipped-DNA repeats: a mechanism of repeat instability. Cell Rep. 37, 110078 (2021).
Zhou, J., Fleming, A. M., Averill, A. M., Burrows, C. J. & Wallace, S. S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 4039–4054 (2015).
Loomis, E. W., Sanz, L. A., Chedin, F. & Hagerman, P. J. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 10, e1004294 (2014).
de Graaff, E. et al. Hotspot for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum. Mol. Genet. 4, 45–49 (1995).
Hayward, B. E. & Usdin, K. Mechanisms of genome instability in the fragile X-related disorders. Genes 12, 1633 (2021).
Rovozzo, R. et al. CGG repeats in the 5′UTR of FMR1 RNA regulate translation of other RNAs localized in the same RNA granules. PLoS ONE 11, e0168204 (2016).
Liu, G. et al. Altered replication in human cells promotes DMPK (CTG)(n). (CAG)(n) repeat instability. Mol. Cell. Biol. 32, 1618–1632 (2012).
Stenson, P. D. et al. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 21, 577–581 (2003).
Kamat, M. A., Bacolla, A., Cooper, D. N. & Chuzhanova, N. A role for non-B DNA forming sequences in mediating microlesions causing human inherited disease. Hum. Mutat. 37, 65–73 (2016).
Weckselblatt, B. & Rudd, M. K. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 31, 587–599 (2015).
Bacolla, A., Tainer, J. A., Vasquez, K. M. & Cooper, D. N. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 44, 5673–5688 (2016).
Raghavan, S. C., Swanson, P. C., Ma, Y. & Lieber, M. R. Double-strand break formation by the RAG complex at the BCL-2 major breakpoint region and at other non-B DNA structures in vitro. Mol. Cell. Biol. 25, 5904–5919 (2005).
Wu, Y. & Brosh, R. M. Jr. G-quadruplex nucleic acids and human disease. FEBS J. 277, 3470–3488 (2010).
Cheloshkina, K. & Poptsova, M. Tissue-specific impact of stem-loops and quadruplexes on cancer breakpoints formation. BMC Cancer 19, 434 (2019).
Cheloshkina, K. & Poptsova, M. Comprehensive analysis of cancer breakpoints reveals signatures of genetic and epigenetic contribution to cancer genome rearrangements. PLoS Comput. Biol. 17, e1008749 (2021).
Kurahashi, H. et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair 5, 1136–1145 (2006).
Di Antonio, M. et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 12, 832–837 (2020).
Del Mundo, I. M. A., Vasquez, K. M. & Wang, G. Modulation of DNA structure formation using small molecules. Biochim. Biophys. Acta 1866, 118539 (2019).
Du, Y. & Zhou, X. Targeting non-B-form DNA in living cells. Chem. Rec. 13, 371–384 (2013).
Vasquez, K. M., Narayanan, L. & Glazer, P. M. Specific mutations induced by triplex-forming oligonucleotides in mice. Science 290, 530–533 (2000).
Nakamori, M. et al. A slipped-CAG DNA-binding small molecule induces trinucleotide-repeat contractions in vivo. Nat. Genet. 52, 146–159 (2020). This study uses a non-B DNA structure-specific ligand to induce repeat contractions at expanded CAG repeats in both cultured human cells and medium spiny neurons of the mouse striatum, suggesting a promising therapeutic approach to reduce pathogenic repeat length.
Dang, D. T., Nguyen, L. T. A., Truong, T. T. T., Nguyen, H. D. & Phan, A. T. Construction of a G-quadruplex-specific DNA endonuclease. Chem. Commun. 57, 4568–4571 (2021).
Zhu, M. et al. Novel roles of an intragenic G-quadruplex in controlling microRNA expression and cardiac function. Nucleic Acids Res. 49, 2522–2536 (2021).
Christensen, L. A., Finch, R. A., Booker, A. J. & Vasquez, K. M. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 66, 4089–4094 (2006).
Boulware, S. B. et al. Triplex-forming oligonucleotides targeting c-MYC potentiate the anti-tumor activity of gemcitabine in a mouse model of human cancer. Mol. Carcinog. 53, 744–752 (2014).
Xu, H. et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 8, 14432 (2017).
Bywater, M. J. et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65 (2012).
Schuldt, A. DNA replication: Pif1 overcomes a quadruplex hurdle. Nat. Rev. Mol. Cell Biol. 12, 402 (2011).
Muellner, J. & Schmidt, K. H. Yeast genome maintenance by the multifunctional PIF1 DNA helicase family. Genes 11, 224 (2020).
Kerrest, A. et al. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat. Struct. Mol. Biol. 16, 159–167 (2009).
Saha, T., Shukla, K., Thakur, R. S., Desingu, A. & Nagaraju, G. Mycobacterium tuberculosis UvrD1 and UvrD2 helicases unwind G-quadruplex DNA. FEBS J. 286, 2062–2086 (2019).
Paul, T. et al. E. coli Rep helicase and RecA recombinase unwind G4 DNA and are important for resistance to G4-stabilizing ligands. Nucleic Acids Res. 48, 6640–6653 (2020).
Eykelenboom, J. K., Blackwood, J. K., Okely, E. & Leach, D. R. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell 29, 644–651 (2008).
Pike, A. C. et al. Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc. Natl Acad. Sci. USA 112, 4286–4291 (2015).
van Wietmarschen, N. et al. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 9, 271 (2018).
Meier, B. et al. Protection of the C. elegans germ cell genome depends on diverse DNA repair pathways during normal proliferation. PLoS ONE 16, e0250291 (2021).
van Wietmarschen, N. et al. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586, 292–298 (2020).
Keller, H. et al. The intrinsically disordered amino-terminal region of human RecQL4: multiple DNA-binding domains confer annealing, strand exchange and G4 DNA binding. Nucleic Acids Res. 42, 12614–12627 (2014).
Budhathoki, J. B. et al. A comparative study of G-quadruplex unfolding and DNA reeling activities of human RECQ5 helicase. Biophys. J. 110, 2585–2596 (2016).
Vannier, J. B., Pavicic-Kaltenbrunner, V., Petalcorin, M. I., Ding, H. & Boulton, S. J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149, 795–806 (2012).
Schult, P. & Paeschke, K. The DEAH helicase DHX36 and its role in G-quadruplex-dependent processes. Biol. Chem. 402, 581–591 (2021).
Jain, A. et al. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 41, 10345–10357 (2013).
Wu, Y., Shin-ya, K. & Brosh, R. M. Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 (2008).
Tarailo-Graovac, M. et al. Spectrum of variations in dog-1/FANCJ and mdf-1/MAD1 defective Caenorhabditis elegans strains after long-term propagation. BMC Genomics 16, 210 (2015).
Kaushik Tiwari, M. & Rogers, F. A. XPD-dependent activation of apoptosis in response to triplex-induced DNA damage. Nucleic Acids Res. 41, 8979–8994 (2013).
Wu, G., Xing, Z., Tran, E. J. & Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl Acad. Sci. USA 116, 20453–20461 (2019).
van Schie, J. J. M. et al. Warsaw breakage syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat. Commun. 11, 4287 (2020).
Wang, Y. et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 10, 943 (2019).
Rich, A. & Zhang, S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 4, 566–572 (2003).
Voloshin, O. N., Mirkin, S. M., Lyamichev, V. I., Belotserkovskii, B. P. & Frank-Kamenetskii, M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature 333, 475–476 (1988).
Frank-Kamenetskii, M. D. & Mirkin, S. M. Triplex DNA structures. Annu. Rev. Biochem. 64, 65–95 (1995).
Lane, A. N., Chaires, J. B., Gray, R. D. & Trent, J. O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 (2008).
Voineagu, I., Narayanan, V., Lobachev, K. S. & Mirkin, S. M. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA 105, 9936–9941 (2008).
Sinden, R. R., Zheng, G. X., Brankamp, R. G. & Allen, K. N. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics 129, 991–1005 (1991).
Castillo-Guzman, D. & Chedin, F. Defining R-loop classes and their contributions to genome instability. DNA Repair 106, 103182 (2021).
Sinden, R. R. DNA Structure and Function (Academic, 1994).
Achaz, G., Coissac, E., Netter, P. & Rocha, E. P. Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics 164, 1279–1289 (2003).
Feschotte, C. & Pritham, E. J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (2007).
Schibler, L. et al. High-resolution comparative mapping among man, cattle and mouse suggests a role for repeat sequences in mammalian genome evolution. BMC Genomics 7, 194 (2006).
Moxon, R., Bayliss, C. & Hood, D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40, 307–333 (2006).
Galen, S. C. et al. Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc. Natl Acad. Sci. USA 112, 13958–13963 (2015).
Lavrov, D. V., Maikova, O. O., Pett, W. & Belikov, S. I. Small inverted repeats drive mitochondrial genome evolution in Lake Baikal sponges. Gene 505, 91–99 (2012).
Deng, Z. et al. A transposon-introduced G-quadruplex motif is selectively retained and constrained to downregulate CYP321A1. Insect Sci. https://doi.org/10.1111/1744-7917.13021 (2022).
Rao, J. E. & Craig, N. L. Selective recognition of pyrimidine motif triplexes by a protein encoded by the bacterial transposon Tn7. J. Mol. Biol. 307, 1161–1170 (2001).
Yin, Y. et al. Molecular mechanisms and topological consequences of drastic chromosomal rearrangements of muntjac deer. Nat. Commun. 12, 6858 (2021).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (2004).
Yuan, J. et al. Simple sequence repeats drive genome plasticity and promote adaptive evolution in penaeid shrimp. Commun. Biol. 4, 186 (2021).
Chan, Y. F. et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010).
Xie, K. T. et al. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363, 81–84 (2019). Structure-specific DSBs and large deletions caused by a Z-DNA-forming GT repeat in the Pel gene from marine stickleback fish populations have important roles in the evolutionary loss of pelvic hindfins in freshwater sticklebacks, suggesting that non-B DNA structure-induced genetic instability has contributed to evolution.
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003).
Acknowledgements
The authors thank A. Wang for support with artwork. The author’s work is supported by the NIH (NIH/NCI CA093729 to K.M.V.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
detectIR: https://sourceforge.net/projects/detectir/
DNA Structure Search: http://utw10685.utweb.utexas.edu/nonbdna/
G4Hunter: http://bioinformatics.ibp.cz
palindrome: http://emboss.bioinformatics.nl/cgi-bin/emboss/palindrome
QGRS Mapper: https://bioinformatics.ramapo.edu/QGRS/index.php
Glossary
- Circular dichroism
-
Absorption spectroscopy method to detect the differential absorption of left- and right-handed light spectra for rapid evaluation of the secondary structures of macromolecules such as protein and DNA.
- DNA helicases
-
A class of motor proteins that move along DNA and transiently separate duplexes into two single strands using energy from ATP hydrolysis.
- Fourier transform infrared spectroscopy
-
A spectroscopy method that simultaneously collects the absorption, emission and photoconductivity of a wide spectral range at high resolution to measure the intensity and wavelength of light required to vibrate molecules in a sample.
- Holliday junctions
-
Branched DNA structures containing four arms covalently linked together that serve as key intermediates in many meiotic and mitotic homologous recombination events.
- Negative supercoiling
-
A segment of underwound DNA in which the two strands wind around the helical axis less than 360° every 10.5 bp and retain twist strain (free energy).
- Okazaki fragments
-
Short fragments of DNA produced by discontinuous replication on the lagging strand during DNA replication. Because the template for lagging strand synthesis is exposed in the 5′–3′ direction at the progressing replication fork, the nascent strand is composed of sequential Okazaki fragments created by DNA polymerase working backwards from the replication fork.
- Satellites
-
A subfraction of genomic DNA consisting of short repetitive nucleotide sequences that are repeated a large number of times. These non-coding repeats are important for centromere and heterochromatin construction and separate from the rest of the genomic DNA on a density gradient because of their higher content of AT base pairs.
- SOS response
-
A complex global response to DNA damage identified in bacteria that includes activation of multiple factors, leading to the stalling of cell division and alteration of DNA replication, recombination and repair to promote genome integrity and cell survival, at the cost of increased mutagenesis.
- Stretching tension
-
When both ends of a segment of DNA are anchored (for example, by proteins) and the DNA is pulled mechanically, it carries stretching tension coupled with twisting torsion along the helix and can be elongated by up to 70% without disrupting base pairs.
- Topoisomerases
-
A class of enzymes that are able to cleave one or both strands of DNA to release topological stress on DNA duplex, and to link or unlink, knot or unknot associated DNA molecules.
- Translesion synthesis polymerases
-
Polymerases that can catalyse DNA polymerization at damaged templates during replication and/or repair, although often with lower fidelty than replicative polymerases.
- Unequal sister chromatid exchange
-
A mitotic crossover event that leads to the exchange of genetic material between homologous chromosomes and is also a major repair pathway for double-strand breaks.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, G., Vasquez, K.M. Dynamic alternative DNA structures in biology and disease. Nat Rev Genet 24, 211–234 (2023). https://doi.org/10.1038/s41576-022-00539-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00539-9
This article is cited by
-
mEnrich-seq: methylation-guided enrichment sequencing of bacterial taxa of interest from microbiome
Nature Methods (2024)
-
AIRE targets poised promoters enriched for Z-DNA
Nature Reviews Genetics (2024)
-
Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats
Nature (2023)
-
G-quadruplexes from non-coding RNAs
Journal of Molecular Medicine (2023)
-
Non-canonical DNA structures in the human ribosomal DNA
Histochemistry and Cell Biology (2023)