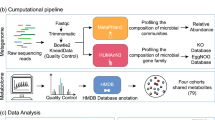
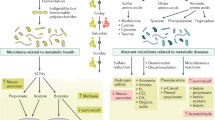
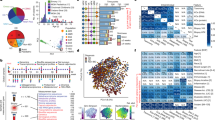
Abstract
The intestine harbours a complex array of microorganisms collectively known as the gut microbiota. The past two decades have witnessed increasing interest in studying the gut microbiota in health and disease, largely driven by rapid innovation in high-throughput multi-omics technologies. As a result, microbial dysbiosis has been linked to many human pathologies, including type 2 diabetes mellitus and inflammatory bowel disease. Integrated analyses of multi-omics data, including metagenomics and metabolomics along with measurements of host response and cataloguing of bacterial isolates, have identified many bacteria and bacterial products that are correlated with disease. Nevertheless, insight into the mechanisms through which microbes affect intestinal health requires going beyond correlation to causation. Current understanding of the contribution of the gut microbiota to disease causality remains limited, largely owing to the heterogeneity of microbial community structures, interindividual differences in disease evolution and incomplete understanding of the mechanisms that integrate microbiota-derived signals into host signalling pathways. In this Review, we provide a broad insight into the microbiome signatures linked to inflammatory and metabolic disorders, discuss outstanding challenges in this field and propose applications of multi-omics technologies that could lead to an improved mechanistic understanding of microorganism–host interactions.
Key points
-
Several commonalities exist between inflammatory bowel disease (IBD) and type 2 diabetes mellitus (T2DM), two multifactorial diseases that show a pattern of increasing global incidence following industrialization.
-
Altered gut bacterial composition and changes in host processing of bacteria-derived metabolites are implicated in IBD and T2DM and provide a shared underlying pathogenetic mechanism.
-
A causal link between dysbiotic microbial communities and IBD or T2DM has been established through gnotobiotic mouse experiments and integrative multi-omics studies.
-
Challenges in disease-specific biomarker discovery include determining the causality of observed changes, understanding their functional redundancy in disease mechanisms and the geographic and ethnic heterogeneity of gut microbiota.
-
Big data refinement, testing and validation of specific bacterial strains, their encoded genes and metabolic byproducts are necessary to identify disease biomarkers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Sender, R., Fuchs, S. & Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340 (2016).
Berg, G. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 103 (2020).
Kaplan, G. G. The global burden of IBD: from 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 12, 720–727 (2015).
Saeedi, P. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 157, 107843 (2019).
Blüher, M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298 (2019).
Chakaroun, R. M., Massier, L. & Kovacs, P. Gut microbiome, intestinal permeability, and tissue bacteria in metabolic disease: perpetrators or bystanders? Nutrients 12, 1082 (2020).
Xue, A. et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 9, 2941 (2018).
de Lange, K. M., Moutsianas, L. & Lee, J. C. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017).
Billings, L. K. & Florez, J. C. The genetics of type 2 diabetes: what have we learned from GWAS? Ann. NY Acad. Sci. 1212, 59–77 (2010).
Yang, S. K. et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63, 80–87 (2014).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Zhou, M. et al. Cause-specific mortality for 240 causes in China during 1990–2013: a systematic subnational analysis for the Global Burden of Disease Study 2013. Lancet 387, 251–272 (2016).
Momozawa, Y. et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun. 9, 2427 (2018).
Qi, Q., Wang, X., Strizich, G. & Wang, T. Genetic determinants of type 2 diabetes in Asians. Int. J. Diabetol. Vasc. Dis. Res. https://doi.org/10.19070/2328-353X-SI01001 (2015).
Yilmaz, B. et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med. 25, 323–336 (2019).
Pascal, V. et al. A microbial signature for Crohn’s disease. Gut 66, 813–822 (2017).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Reitmeier, S. et al. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28, 258–272.e6 (2020).
Metwaly, A. et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 11, 4322 (2020).
Ananthakrishnan, A. N. et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 21, 603–610.e3 (2017).
Round, J. L. & Mazmanian, S. K. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Jurjus, A. et al. Inflammatory bowel disease, colorectal cancer and type 2 diabetes mellitus: the links. BBA Clin. 5, 16–24 (2015).
Verdugo-Meza, A., Ye, J., Dadlani, H., Ghosh, S. & Gibson, D. L. Connecting the dots between inflammatory bowel disease and metabolic syndrome: a focus on gut-derived metabolites. Nutrients 12, 1434 (2020).
Kootte, R. S. et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 26, 611–619.e6 (2017).
Wang, H. et al. Promising treatment for type 2 diabetes: fecal microbiota transplantation reverses insulin resistance and impaired islets. Front. Cell. Infect. Microbiol. 9, 455 (2020).
Sokol, H. et al. Fecal microbiota transplantation to maintain remission in Crohn’s disease: a pilot randomized controlled study. Microbiome 8, 12 (2020).
Costello, S. P. et al. Systematic review with meta-analysis: faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment. Pharmacol. Ther. 46, 213–224 (2017).
Vrieze, A. et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7 (2012).
Paramsothy, S., Kamm, M. & Kaakoush, N. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017).
Gupta, A. & Khanna, S. Fecal microbiota transplantation. JAMA 318, 102 (2017).
Quraishi, M. N. et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment. Pharmacol. Ther. 46, 479–493 (2017).
Mocanu, V. et al. Fecal microbial transplantation and fiber supplementation in patients with severe obesity and metabolic syndrome: a randomized double-blind, placebo-controlled phase 2 trial. Nat. Med. 27, 1272–1279 (2021).
van Lier, Y. F. et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci. Transl Med. 12, eaaz8926 (2020).
Rossen, N. G. et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118.e4 (2015).
Moayyedi, P. et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6 (2015).
Sood, A. et al. Acceptability, tolerability, and safety of fecal microbiota transplantation in patients with active ulcerative colitis (AT&S Study). J. Gastroenterol. Hepatol. 35, 418–424 (2020).
Xiang, L. et al. Efficacy of faecal microbiota transplantation in Crohn’s disease: a new target treatment? Microb. Biotechnol. 13, 760–769 (2020).
de Groot, P. et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69, 502–512 (2020).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2017).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Schirmer, M. et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 24, 600–610.e4 (2018).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014).
Sokol, H. & Seksik, P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr. Opin. Gastroenterol. 26, 327–331 (2010).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79–R79 (2012).
Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500 (2015).
Vila, A. V. et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl Med. 10, eaap8914 (2019).
Schirmer, M., Garner, A., Vlamakis, H. & Xavier, R. J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 17, 497–511 (2019).
D’Haens, G. R. et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 114, 262–267 (1998).
Rutgeerts, P. et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet 338, 771–774 (1991).
Perencevich, M. & Burakoff, R. Use of antibiotics in the treatment of inflammatory bowel disease. Inflamm. Bowel Dis. 12, 651–664 (2006).
Hörmannsperger, G., Schaubeck, M. & Haller, D. Intestinal microbiota in animal models of inflammatory diseases. ILAR J. 56, 179–191 (2015).
Ahmed, M., Metwaly, A. & Haller, D. Modeling microbe-host interaction in the pathogenesis of Crohn’s disease. Int. J. Med. Microbiol. 311, 151489 (2021).
Nagao-Kitamoto, H. et al. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2, 468–481 (2016).
Sellon, R. K. et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66, 5224–5231 (1998).
Schaubeck, M. et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 65, 225–237 (2016).
Britton, G. J. et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity 50, 212–224.e4 (2019).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
He, Y. et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24, 1532–1535 (2018).
Zhong, H. et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naive type 2 diabetics. EBioMedicine 47, 373–383 (2019).
Karlsson, F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Shin, N.-R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014).
Org, E. et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569 (2015).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071 (2013).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Gilijamse, P. W. et al. Treatment with Anaerobutyricum soehngenii: a pilot study of safety and dose–response effects on glucose metabolism in human subjects with metabolic syndrome. NPJ Biofilms Microbiomes 6, 16 (2020).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Zhou, W. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569, 663–671 (2019).
Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95 (2001).
Califf, R. M. Biomarker definitions and their applications. Exp. Biol. Med. 243, 213–221 (2018).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004).
Joossens, M. et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637 (2011).
Fang, X. et al. Metagenomics-based, strain-level analysis of Escherichia coli from a time-series of microbiome samples from a Crohn’s disease patient. Front. Microbiol. 9, 2559 (2018).
Hall, A. B. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J. & Martinez-Medina, M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 11, 841–852 (2017).
Lopez-Siles, M. et al. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int. J. Med. Microbiol. 304, 464–475 (2014).
Chehoud, C. et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 21, 1948–1956 (2015).
Sarrabayrouse, G. et al. Fungal and bacterial loads: noninvasive inflammatory bowel disease biomarkers for the clinical setting. mSystems 6, e01277-20 (2021).
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2, 17004 (2017).
Martínez, I. et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 11, 527–538 (2015).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Blaser, M. J. & Falkow, S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894 (2009).
Clooney, A. G. et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut 70, 499–510 (2021).
Vangay, P. et al. US immigration westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018).
He, Y. et al. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome 6, 172 (2018).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Thingholm, L. B. et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 26, 252–264.e10 (2019).
Zhang, F. et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 66, 485–493 (2019).
Chávez-Carbajal, A. et al. Characterization of the gut microbiota of individuals at different T2D stages reveals a complex relationship with the host. Microorganisms 8, 94 (2020).
Li, Q. et al. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 10, 5450 (2020).
Ahmad, A. et al. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLoS ONE 14, e0226372 (2020).
Lopez-Siles, M. et al. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 78, 420–428 (2012).
Duncan, S. H., Hold, G. L., Barcenilla, A., Stewart, C. S. & Flint, H. J. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52, 1615–1620 (2002).
Moossavi, S. The necessity for an Iranian gut microbiome initiative. Middle East J. Dig. Dis. 6, 109–110 (2014).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Franzosa, E. A. et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305 (2019).
Jacobs, J. P. et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2, 750–766 (2016).
Kolho, K.-L., Pessia, A., Jaakkola, T., de Vos, W. M. & Velagapudi, V. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J. Crohns Colitis 11, 321–334 (2017).
Guasch-Ferré, M. et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care 39, 833–846 (2016).
Tam, Z. Y. et al. Metabolite profiling in identifying metabolic biomarkers in older people with late-onset type 2 diabetes mellitus. Sci. Rep. 7, 4392 (2017).
Urpi-Sarda, M. et al. Metabolomics for biomarkers of type 2 diabetes mellitus: advances and nutritional intervention trends. Curr. Cardiovasc. Risk Rep. 9, 12 (2015).
Preter, V. D. et al. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut 64, 447–458 (2015).
Santoru, M. L. et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 7, 9523 (2017).
Ni, J. et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Transl Med. 9, eaah6888 (2017).
Marchesi, J. R. et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 6, 546–551 (2007).
Jansson, J. et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE 4, e6386 (2009).
Le Gall, G. et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 10, 4208–4218 (2011).
Bain, J. R. et al. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes 58, 2429–2443 (2009).
Menni, C. et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62, 4270–4276 (2013).
Batch, B. C. et al. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism 62, 961–969 (2013).
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 (2011).
Würtz, P. et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 36, 648–655 (2013).
Salihovic, S. et al. Non-targeted urine metabolomics and associations with prevalent and incident type 2 diabetes. Sci. Rep. 10, 16474 (2020).
Wang-Sattler, R. et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 8, 615 (2012).
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016).
Nikolaus, S. et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 153, 1504–1516.e2 (2017).
Chen, T. et al. Tryptophan predicts the risk for future type 2 diabetes. PLoS ONE 11, e0162192 (2016).
Hisamatsu, T. et al. Novel, objective, multivariate biomarkers composed of plasma amino acid profiles for the diagnosis and assessment of inflammatory bowel disease. PLoS ONE 7, e31131 (2012).
Roager, H. M. & Licht, T. R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 9, 3294 (2018).
Lavelle, A. & Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237 (2020).
Sorgdrager, F. J. H., Naudé, P. J. W., Kema, I. P., Nollen, E. A. & De Deyn, P. P. Tryptophan metabolism in inflammaging: from biomarker to therapeutic target. Front. Immunol. 10, 2565 (2019).
Tillin, T. et al. Diabetes risk and amino acid profiles: cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall And Brent Revisited) Study. Diabetologia 58, 968–979 (2015).
Schoepfer, A. M. et al. Fecal calprotectin correlates more closely with the simple endoscopic score for Crohn’s disease (SES-CD) than CRP, blood leukocytes, and the CDAI. Am. J. Gastroenterol. 105, 162–169 (2010).
Lewis, J. D. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology 140, 1817–1826.e2 (2011).
Sipponen, T. & Kolho, K.-L. Fecal calprotectin in diagnosis and clinical assessment of inflammatory bowel disease. Scand. J. Gastroenterol. 50, 74–80 (2015).
Vich Vila, A. et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl Med. 10, eaap8914 (2018).
Dubinsky, V. et al. Dysbiosis in metabolic genes of the gut microbiomes of patients with an ileo-anal pouch resembles that observed in Crohn’s disease. mSystems 6, e00984-20 (2021).
Long, J. et al. Metabolite biomarkers of type 2 diabetes mellitus and pre-diabetes: a systematic review and meta-analysis. BMC Endocr. Disord. 20, 174 (2020).
Wu, H. et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 32, 379–390.e3 (2020).
Floegel, A. et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 62, 639–648 (2013).
Lu, Y. et al. Metabolic signatures and risk of type 2 diabetes in a Chinese population: an untargeted metabolomics study using both LC-MS and GC-MS. Diabetologia 59, 2349–2359 (2016).
Buttó, L. F., Schaubeck, M. & Haller, D. Mechanisms of microbe-host interaction in Crohn’s disease: dysbiosis vs. pathobiont selection. Front. Immunol. 6, 555 (2015).
Jochum, L. & Stecher, B. Label or concept — what is a pathobiont? Trends Microbiol. 28, 789–792 (2020).
Buttó, L. F. & Haller, D. Dysbiosis in Crohn’s disease — joint action of stochastic injuries and focal inflammation in the gut. Gut Microbes 8, 53–58 (2017).
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Arrieta, M., Sadarangani, M., Brown, E. M., Russell, S. L. & Nimmo, M. A humanized microbiota mouse model of ovalbumin-induced lung inflammation. Gut Microbes 7, 342–352 (2016).
Blanton, L.V. et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016).
Kostic, A. D. et al. The dynamics of the human infant gut microbiome in development and in progression towards type 1 diabetes. Cell Host Microbe 17, 260–273 (2016).
Steck, N. et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971 (2011).
Lengfelder, I. et al. Complex bacterial consortia reprogram the colitogenic activity of Enterococcus faecalis in a gnotobiotic mouse model of chronic, immune-mediated colitis. Front. Immunol. 10, 1420 (2019).
Kim, S. C., Tonkonogy, S. L., Karrasch, T., Jobin, C. & Balfour Sartor, R. Dual-association of gnotobiotic IL-10–/– mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm. Bowel Dis. 13, 1457–1466 (2007).
Eun, C. S. et al. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10–/– mice. Infect. Immun. 82, 2239–2246 (2014).
Brugiroux, S. et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar typhimurium. Nat. Microbiol. 2, 16215 (2016).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Rehman, A. et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 65, 238–248 (2016).
Turnbaugh, P. J. & Gordon, J. I. The core gut microbiome, energy balance and obesity. J. Physiol. 587, 4153–4158 (2009).
Walter, J., Armet, A. M., Finlay, B. B. & Shanahan, F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232 (2020).
Brown, K. et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 10, 321–332 (2016).
Cani, P. D. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 (2007).
Vester-Andersen, M. K. et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci. Rep. 9, 13473 (2019).
Sanna, S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605 (2019).
Kishikawa, T. et al. A metagenome-wide association study of gut microbiome in patients with multiple sclerosis revealed novel disease pathology. Front. Cell. Infect. Microbiol. 10, 585973 (2020).
Doumatey, A. P. et al. Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front. Cell. Infect. Microbiol. 10, 63 (2020).
Peters, B. A. et al. A taxonomic signature of obesity in a large study of American adults. Sci. Rep. 8, 9749 (2018).
Acknowledgements
The authors’ research is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project number 395357507 (SFB 1371, Microbiome Signatures) — and has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement number 964590.
Author information
Authors and Affiliations
Contributions
A.M. and D.H. contributed equally to all aspects of the article. In addition, S.R. researched data for the article, contributed substantially to discussion of its content, and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Herbert Tilg, Chaysavanh Manichanh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Microbiome
-
All the microorganisms, their genomes and the surrounding host-shaped environmental conditions of a given habitat. The gut microbiome can be characterized through the application of metagenomics, metabolomics, metatranscriptomics and metaproteomics combined with clinical or environmental metadata.
- Microbiota
-
The microorganisms (bacteria, fungi, archaea and viruses) of a defined environment.
- Microbiome signatures
-
Unique patterns of microbiome configuration that can stratify defined physiological and pathological conditions and can be used to predict the risk of disease development or progression.
- Pathobionts
-
Microorganisms that are linked to chronic inflammatory conditions but are harmless to the host under normal conditions.
- Faecal microbiota transplantation
-
(FMT). A treatment based on transfer of microorganisms from the stool of a healthy donor into a patient’s digestive system.
- Dysbiosis
-
(Also known as pathobiome). An altered microbial community composition that affects the host immune response and leads to the emergence and outbreak of pathogens.
- Specificity
-
The proportion of individuals who genuinely do not have the disease or condition and receive negative test results.
- Sensitivity
-
The proportion of individuals who genuinely have the disease or condition and receive positive test results.
- Diagnostic biomarkers
-
Characteristics that are directly linked to the aetiology of the disease such as elevated blood glucose concentration (which is diagnostic of type 2 diabetes mellitus) and markers that show a strong correlation with inflammation in inflammatory bowel disease (such as faecal calprotectin levels).
Rights and permissions
About this article
Cite this article
Metwaly, A., Reitmeier, S. & Haller, D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat Rev Gastroenterol Hepatol 19, 383–397 (2022). https://doi.org/10.1038/s41575-022-00581-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-022-00581-2
This article is cited by
-
Multiomics-based molecular subtyping based on the commensal microbiome predicts molecular characteristics and the therapeutic response in breast cancer
Molecular Cancer (2024)
-
Evolutionarily related host and microbial pathways regulate fat desaturation in C. elegans
Nature Communications (2024)
-
Effects of water ammonia nitrogen on hemolymph and intestinal microbiota of Litopenaeus vannamei
Advanced Biotechnology (2024)
-
New dawn of ginsenosides: regulating gut microbiota to treat metabolic syndrome
Phytochemistry Reviews (2024)
-
Metabolic and inflammatory perturbation of diabetes associated gut dysbiosis in people living with and without HIV infection
Genome Medicine (2024)