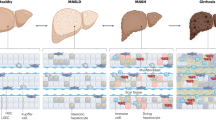
Abstract
Macrophages have diverse phenotypes and functions due to differences in their origin, location and pathophysiological context. Although their main role in the liver has been described as immunoregulatory and detoxifying, changes in macrophage phenotypes, diversity, dynamics and function have been reported during obesity-related complications such as non-alcoholic fatty liver disease (NAFLD). NAFLD encompasses multiple disease states from hepatic steatosis to non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis and hepatocarcinoma. Obesity and insulin resistance are prominent risk factors for NASH, a disease with a high worldwide prevalence and no approved treatment. In this Review, we discuss the turnover and function of liver-resident macrophages (Kupffer cells) and monocyte-derived hepatic macrophages. We examine these populations in both steady state and during NAFLD, with an emphasis on NASH. The explosion in high-throughput gene expression analysis using single-cell RNA sequencing (scRNA-seq) within the last 5 years has revolutionized the study of macrophage heterogeneity, substantially increasing our understanding of the composition and diversity of tissue macrophages, including in the liver. Here, we highlight scRNA-seq findings from the last 5 years on the diversity of liver macrophages in homeostasis and metabolic disease, and reveal hepatic macrophage function beyond their classically described inflammatory role in the progression of NAFLD and NASH pathogenesis.
Key points
-
Macrophages are highly plastic cells of the immune system that can acquire a spectrum of phenotypes according to their spatiotemporal pathophysiological context.
-
Liver macrophages are either embryo-derived resident macrophages or recruited peripheral monocyte-derived macrophages.
-
Liver macrophages have been shown to contribute to non-alcoholic fatty liver disease (NAFLD) progression in obesity through production of both inflammatory and non-inflammatory factors.
-
Single-cell RNA sequencing (scRNA-seq) has identified distinct liver macrophage subsets in mice and humans in health and liver disease.
-
scRNA-seq has enabled the identification of novel pathogenic factors expressed by liver macrophages that could exacerbate or protect from NAFLD progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gordon, S. & Plüddemann, A. Tissue macrophages: heterogeneity and functions. BMC Biol. 15, 53–53 (2017).
Liddiard, K. & Taylor, P. R. Understanding local macrophage phenotypes in disease: shape-shifting macrophages. Nat. Med. 21, 119–120 (2015).
Pollard, J. W. Trophic macrophages in development and disease. Nat. Rev. Immunol. 9, 259–270 (2009).
Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017).
Wen, Y., Lambrecht, J., Ju, C. & Tacke, F. Hepatic macrophages in liver homeostasis and diseases–diversity, plasticity and therapeutic opportunities. Cell. Mol. Immunol. 18, 45–56 (2021).
Kazankov, K. et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 16, 145–159 (2019).
David, B. A. et al. Combination of mass cytometry and imaging analysis reveals origin, location, and functional repopulation of liver myeloid cells in mice. Gastroenterology 151, 1176–1191 (2016).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
Bonnardel, J. et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51, 638–654.e9 (2019).
Crispe, I. N. Liver antigen-presenting cells. J. Hepatol. 54, 357–365 (2011).
Sierro, F. et al. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity 47, 374–388.e6 (2017).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e19 (2019).
Seidman, J. S. et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 52, 1057–1074.e7 (2020). This paper characterized NASH-specific transcriptional programmes driving the phenotype of liver macrophage populations during metabolic disease.
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Soucie, E. L. et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351, aad5510 (2016).
Bleriot, C. et al. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Geissmann, F., Jung, S. & Littman, D. R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003).
Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B. L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Sunderkotter, C. et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J. Immunol. 172, 4410–4417 (2004).
van de Laar, L. et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44, 755–768 (2016).
Sakai, M. et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity 51, 655–670.e8 (2019).
Devisscher, L. et al. Non-alcoholic steatohepatitis induces transient changes within the liver macrophage pool. Cell. Immunol. 322, 74–83 (2017).
Zigmond, E. et al. Infiltrating monocyte-derived macrophages and resident Kupffer cells display different ontogeny and functions in acute liver injury. J. Immunol. 193, 344–353 (2014).
Wang, J. & Kubes, P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 165, 668–678 (2016).
Jin, H. et al. Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury. Nat. Commun. 12, 2863 (2021).
Ingersoll, M. A. et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115, e10–e19 (2010).
Passlick, B., Flieger, D. & Ziegler-Heitbrock, H. W. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74, 2527–2534 (1989).
Patel, A. A. et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 214, 1913–1923 (2017).
Tak, T. et al. Circulatory and maturation kinetics of human monocyte subsets in vivo. Blood 130, 1474–1477 (2017).
Eguíluz-Gracia, I. M. D. et al. Rapid recruitment of CD14+ monocytes in experimentally induced allergic rhinitis in human subjects. J. Allergy Clin. Immunol. 137, 1872–1881.e12 (2016).
Jardine, L. et al. Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nat. Commun. 10, 1999 (2019).
van der Laan, A. M. et al. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur. Heart J. 35, 376–385 (2014).
Bittmann, I. et al. The role of graft-resident Kupffer cells and lymphocytes of donor type during the time course after liver transplantation–a clinico-pathological study. Virchows Arch. 443, 541–548 (2003).
Pallett, L. J. et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J. Exp. Med. 217, e20200050 (2020). This paper studied the turnover of human liver macrophages in patients undergoing liver transplantation.
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77–77 (2016).
Mereu, E. et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755 (2020).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 (2017).
Brancale, J. & Vilarinho, S. A single cell gene expression atlas of 28 human livers. J. Hepatol. 75, 219–220 (2021).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018). This paper characterized two distinct intrahepatic populations of liver macrophages in healthy human livers.
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). This paper characterized the heterogeneity of human liver macrophages in healthy and cirrhotic livers and identified two pathogenic macrophage populations associated with liver disease.
Zhao, J. et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 6, 22–22 (2020).
Wu, X. et al. Human liver macrophage subsets defined by CD32. Front. Immunol. 11, 2108 (2020).
Aizarani, N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019).
Andrews, T. S. et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol. Commun. 6, 821–840 (2021).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Krenkel, O. et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut 69, 551–563 (2019).
Remmerie, A. et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 53, 641–657.e14 (2020). This paper characterized a distinct subset of recruited macrophages during the progression of NASH.
Scott, C. L. et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 49, 312–325.e5 (2018).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660.e5 (2019).
Blériot, C. et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity 54, 2101–2116.e6 (2021). This paper identified two functionally distinct populations of embryo-derived Kuppfer cells in mice.
De Simone, G. et al. Identification of a Kupffer cell subset capable of reverting the T cell dysfunction induced by hepatocellular priming. Immunity 54, 2089–2100.e8 (2021).
Tran, S. et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity 53, 627–640.e5 (2020). This paper demonstrated Kupffer cell renewal is impaired in NASH, leading to increased recruitment of monocyte-derived macrophages with altered metabolic responses to liver disease.
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Bykov, I., Ylipaasto, P., Eerola, L. & Lindros, K. O. Functional differences between periportal and perivenous Kupffer cells isolated by digitonin-collagenase perfusion. Comp. Hepatol. 3 (Suppl. 1), 34 (2004).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Daemen, S. et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 34, 108626 (2021).
Neuschwander-Tetri, B. A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology 52, 774–788 (2010).
Younossi, Z. M. et al. Global epidemiology of nonalcoholic fatty liver disease–meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016).
Albhaisi, S., Chowdhury, A. & Sanyal, A. J. Non-alcoholic fatty liver disease in lean individuals. JHEP Rep. 1, 329–341 (2019).
Younossi, Z. M. et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J. Hepatol. 71, 793–801 (2019).
Hardy, T., Oakley, F., Anstee, Q. M. & Day, C. P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu. Rev. Pathol. Mech. Dis. 11, 451–496 (2016).
Feldstein, A. E. et al. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology 125, 437–443 (2003).
Ribeiro, P. S. et al. Hepatocyte apoptosis, expression of death receptors, and activation of NF-κB in the liver of nonalcoholic and alcoholic steatohepatitis patients. Am. J. Gastroenterol. 99, 1708–1717 (2004).
Alonso, C. et al. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology 152, 1449–1461.e7 (2017).
Tilg, H. & Moschen, A. R. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 52, 1836–1846 (2010).
Tamura, S. & Shimomura, I. Contribution of adipose tissue and de novo lipogenesis to nonalcoholic fatty liver disease. J. Clin. Invest. 115, 1139–1142 (2005).
Marchesini, G. et al. Association of nonalcoholic fatty liver disease with insulin resistance. Am. J. Med. 107, 450–455 (1999).
Hirsova, P., Ibrahim, S. H., Gores, G. J. & Malhi, H. Lipotoxic lethal and sublethal stress signaling in hepatocytes: relevance to NASH pathogenesis. J. Lipid Res. 57, 1758–1770 (2016).
Puri, P. et al. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology 134, 568–576 (2008).
Mota, M., Banini, B. A., Cazanave, S. C. & Sanyal, A. J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism 65, 1049–1061 (2016).
Huang, W. et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 59, 347–357 (2010).
Neyrinck, A. M. et al. Critical role of Kupffer cells in the management of diet-induced diabetes and obesity. Biochem. Biophys. Res. Commun. 385, 351–356 (2009).
Thomas, D. & Apovian, C. Macrophage functions in lean and obese adipose tissue. Metab. Clin. Exp. 72, 120–143 (2017).
Myoung Sook, H. et al. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218–222 (2013).
Cai, D. et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-β and NF-κB. Nat. Med. 11, 183–190 (2005).
Obstfeld, A. E. et al. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes 59, 916–925 (2010).
Stienstra, R. et al. Kupffer cells promote hepatic steatosis via interleukin-1β-dependent suppression of peroxisome proliferator-activated receptor α activity. Hepatology 51, 511–522 (2010).
Papackova, Z. et al. Kupffer cells ameliorate hepatic insulin resistance induced by high-fat diet rich in monounsaturated fatty acids: the evidence for the involvement of alternatively activated macrophages. Nutr. Metab. 9, 22 (2012).
Lanthier, N. et al. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G107–G116 (2010).
Clementi, A. H., Gaudy, A. M., van Rooijen, N., Pierce, R. H. & Mooney, R. A. Loss of Kupffer cells in diet-induced obesity is associated with increased hepatic steatosis, STAT3 signaling, and further decreases in insulin signaling. Biochim. Biophys. Acta 1792, 1062–1072 (2009).
Chen, L. et al. Selective depletion of hepatic Kupffer cells significantly alleviated hepatosteatosis and intrahepatic inflammation induced by high fat diet. Hepatogastroenterology 59, 1208–1212 (2012).
Odegaard, J. I. et al. Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab. 7, 496–507 (2008).
Maeda, S. et al. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 11, 191–198 (2005).
Morinaga, H. et al. Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice. Diabetes 64, 1120–1130 (2015).
Morgantini, C. et al. Liver macrophages regulate systemic metabolism through non-inflammatory factors. Nat. Metab. 1, 445–459 (2019).
Ramachandran, P., Kylie, K. P., Dobie, R., Wilson-Kanamori, J. R. & Henderson, N. C. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 17, 457–472 (2020).
Tencerova, M. et al. Activated Kupffer cells inhibit insulin sensitivity in obese mice. FASEB J. 29, 2959–2969 (2015).
Azzimato, V. et al. Liver macrophages inhibit the endogenous antioxidant response in obesity-associated insulin resistance. Sci. Transl. Med. 12, eaaw9709 (2020).
Aouadi, M. et al. Lipid storage by adipose tissue macrophages regulates systemic glucose tolerance. Am. J. Physiol. Endocrinol. Metab. 307, E374–E383 (2014).
Kosteli, A. et al. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest. 120, 3466–3479 (2010).
Wan, J. et al. M2 Kupffer cells promote M1 Kupffer cell apoptosis: a protective mechanism against alcoholic and nonalcoholic fatty liver disease. Hepatology 59, 130–142 (2014).
Ramachandran, P. et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl Acad. Sci. USA 109, E3186–E3195 (2012).
Issa, R. et al. Apoptosis of hepatic stellate cells: involvement in resolution of biliary fibrosis and regulation by soluble growth factors. Gut 48, 548–557 (2001).
Morita, Y. et al. Impact of tissue macrophage proliferation on peripheral and systemic insulin resistance in obese mice with diabetes. BMJ Open Diabetes Res. Care 8, e001578 (2020).
Karlmark, K. R. et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50, 261–274 (2009).
Seki, E. et al. CCR2 promotes hepatic fibrosis in mice. Hepatology 50, 185–197 (2009).
Zimmermann, H. W. et al. Functional contribution of elevated circulating and hepatic non-classical CD14CD16 monocytes to inflammation and human liver fibrosis. PLoS ONE 5, e11049 (2010).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/record/NCT03028740 (2022).
Francque, S. et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat. Rev. Gastroenterol. Hepatol. 18, 24–39 (2021).
Francque, S. M. et al. A randomized, controlled trial of the pan-PPAR agonist lanifibranor in NASH. N. Engl. J. Med. 385, 1547–1558 (2021).
Deczkowska, A. et al. XCR1+ type 1 conventional dendritic cells drive liver pathology in non-alcoholic steatohepatitis. Nat. Med. 27, 1043–1054 (2021).
Bril, F. et al. Metabolic and histological implications of intrahepatic triglyceride content in nonalcoholic fatty liver disease. Hepatology 65, 1132–1144 (2017).
Rosso, C. et al. Crosstalk between adipose tissue insulin resistance and liver macrophages in non-alcoholic fatty liver disease. J. Hepatol. 71, 1012–1021 (2019).
Lomonaco, R. et al. Metabolic impact of nonalcoholic steatohepatitis in obese patients with type 2 diabetes. Diabetes Care 39, 632–638 (2016).
Priest, C. & Tontonoz, P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 1, 1177–1188 (2019).
Metlakunta, A. et al. Kupffer cells facilitate the acute effects of leptin on hepatic lipid metabolism. Am. J. Physiol. 312, E11–E18 (2017).
Azzimato, V. et al. Hepatic miR-144 drives fumarase activity preventing NRF2 activation during obesity. Gastroenterology 161, 1982–1997.e11 (2021).
Li, H. et al. Crosstalk between liver macrophages and surrounding cells in nonalcoholic steatohepatitis. Front. Immunol. 11, 1169 (2020).
Chu, P. S. et al. C-C motif chemokine receptor 9 positive macrophages activate hepatic stellate cells and promote liver fibrosis in mice. Hepatology 58, 337–350 (2013).
Pradere, J. P. et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 58, 1461–1473 (2013).
Thomas, J. A. et al. Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function. Hepatology 53, 2003–2015 (2011).
Kremer, M. et al. Kupffer cell and interleukin-12-dependent loss of natural killer T cells in hepatosteatosis. Hepatology 51, 130–141 (2010).
Hirsova, P. et al. Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 150, 956–967 (2016).
Tateya, S. et al. Endothelial NO/cGMP/VASP signaling attenuates Kupffer cell activation and hepatic insulin resistance induced by high-fat feeding. Diabetes 60, 2792–2801 (2011).
McMahan, R. H., Porsche, C. E., Edwards, M. G. & Rosen, H. R. Free fatty acids differentially downregulate chemokines in liver sinusoidal endothelial cells: insights into non-alcoholic fatty liver disease. PLoS ONE 11, e0159217 (2016).
Liu, X. L. et al. Lipotoxic hepatocyte-derived exosomal microRNA 192-5p activates macrophages through Rictor/Akt/forkhead box transcription factor O1 signaling in nonalcoholic fatty liver disease. Hepatology 72, 454–469 (2020).
Miyachi, Y. et al. Roles for cell-cell adhesion and contact in obesity-induced hepatic myeloid cell accumulation and glucose intolerance. Cell Rep. 18, 2766–2779 (2017).
Al Attar, A., Antaramian, A. & Noureddin, M. Review of galectin-3 inhibitors in the treatment of nonalcoholic steatohepatitis. Expert. Rev. Clin. Pharmacol. 14, 457–464 (2021).
Acknowledgements
The authors thank R. Harris (Department of Clinical Neuroscience, Karolinska Institutet, Center for Molecular Medicine, Karolinska University Hospital, 171 76 Stockholm, Sweden) and S. Craige (Department of Human Nutrition, Foods, and Exercise, Virginia Tech, Blacksburg, Virginia) for their valuable comments on the manuscript. M.A., E.B. and P.C. acknowledge the support of funds from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 864788), the EFSD supported by EFDS/Lilly European Diabetes research programme, the Karolinska Institutet, the Swedish Research Council (M.A.; 2015-03582 and 2019-01056), the Novo Nordisk Foundation (M.A.; NNF20OC0060053, NNF19OC0057127), including the Metabolite-Related Inflammation and Disease Consortium (MeRIAD; NNF0064142), and the Strategic Research Programmes in Diabetes (M.A.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Endocrinology thanks Charlotte Scott, Frank Tacke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Fate mapping
-
Labelling of specific cell subsets in the embryo to trace their contribution to cell populations and tissues in the adult organism.
- Yolk sac
-
A sac attached to the embryo during development that provides nutrients and cells to the embryo.
- Sterile injury
-
Injury or inflammation not caused by pathogenic infection.
- Tissue monocytes
-
Cell population identified in the liver tissue by single-cell RNA sequencing (scRNA-seq) with a phenotype similar to circulating monocytes.
- Scar-associated macrophages
-
Liver macrophages with a lipid-handling phenotype mainly associated with the development of liver fibrosis in patients with cirrhosis.
- Tissue-specific enhancer
-
A regulatory element that induces tissue-specific transcription factors shaping the phenotype of tissue-resident cells.
- Lipid-associated macrophages
-
Tissue macrophages with a lipid handling phenotype mainly associated with the development of obesity and non-alcoholic steatohepatitis.
- Crown-like structures
-
Histological finding where macrophages are surrounding lipid-laden hepatocytes (in the liver) or dying adipocytes (in the adipose tissue) in a crown-like structure.
- Liver non-parenchymal cells
-
Liver cells that do not make up the liver parenchyma including liver sinusoidal endothelial cells, hepatic stellate cells and immune cells.
- Temporal trajectory analysis
-
In silico analysis of cell trajectories or cell differentiation patterns based on scRNA-seq data.
Rights and permissions
About this article
Cite this article
Barreby, E., Chen, P. & Aouadi, M. Macrophage functional diversity in NAFLD — more than inflammation. Nat Rev Endocrinol 18, 461–472 (2022). https://doi.org/10.1038/s41574-022-00675-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41574-022-00675-6
This article is cited by
-
Macrophage-specific FGFR1 deletion alleviates high-fat-diet-induced liver inflammation by inhibiting the MAPKs/TNF pathways
Acta Pharmacologica Sinica (2024)
-
The implications of FASN in immune cell biology and related diseases
Cell Death & Disease (2024)
-
The dual function of cGAS-STING signaling axis in liver diseases
Acta Pharmacologica Sinica (2024)
-
Spatially-resolved transcriptomics reveal macrophage heterogeneity and prognostic significance in diffuse large B-cell lymphoma
Nature Communications (2024)
-
Butyrate limits inflammatory macrophage niche in NASH
Cell Death & Disease (2023)