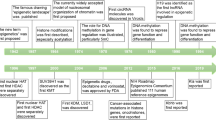
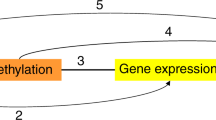
Abstract
Pioneering studies performed over the past few decades demonstrate links between epigenetics and type 2 diabetes mellitus (T2DM), the metabolic disorder with the most rapidly increasing prevalence in the world. Importantly, these studies identified epigenetic modifications, including altered DNA methylation, in pancreatic islets, adipose tissue, skeletal muscle and the liver from individuals with T2DM. As non-genetic factors that affect the risk of T2DM, such as obesity, unhealthy diet, physical inactivity, ageing and the intrauterine environment, have been associated with epigenetic modifications in healthy individuals, epigenetics probably also contributes to T2DM development. In addition, genetic factors associated with T2DM and obesity affect the epigenome in human tissues. Notably, causal mediation analyses found DNA methylation to be a potential mediator of genetic associations with metabolic traits and disease. In the past few years, translational studies have identified blood-based epigenetic markers that might be further developed and used for precision medicine to help patients with T2DM receive optimal therapy and to identify patients at risk of complications. This Review focuses on epigenetic mechanisms in the development of T2DM and the regulation of body weight in humans, with a special focus on precision medicine.
Key points
-
Type 2 diabetes mellitus (T2DM) is a common metabolic disease that involves insulin resistance of adipose tissue, skeletal muscle and the liver, and impaired insulin secretion from the pancreatic islets.
-
As a multifactorial disease, T2DM has a genetic component; epigenetics might explain the missing heritability, and it is also strongly influenced by non-genetic factors, for example, exercise, diet, obesity and ageing.
-
Epigenetic alterations, including DNA methylation, are found in several tissues from patients with T2DM and are also associated with environmental factors known to increase risk of the disease.
-
Human studies demonstrate how the epigenome mediates the effects of both genetic variation and environmental exposures on gene expression and cell functions associated with T2DM and obesity.
-
Epigenetic factors might be induced by the environment, heritable, or stochastic, and they act together to define the epigenetic profile of each cell and tissue.
-
Epigenetic marks can potentially be used as biomarkers for prediction of T2DM, risk of vascular complications, and response to therapy and lifestyle interventions, thereby offering a tool for precision medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Volkov, P. et al. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes 66, 1074–1085 (2017).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012).
Eliasson, L. & Regazzi, R. Micro(RNA) management and mismanagement of the islet. J. Mol. Biol. 432, 1419–1428 (2020).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Da Silva Xavier, G. The cells of the islets of Langerhans. J. Clin. Med. 7, 54 (2018).
Galicia-Garcia, U. et al. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 21, 6275 (2020).
Avrahami, D. et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metab. 22, 619–632 (2015).
Bacos, K. et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat. Commun. 7, 11089 (2016).
Daneshpajooh, M. et al. HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia 60, 116–125 (2017).
Daneshpajooh, M., Eliasson, L., Bacos, K. & Ling, C. MC1568 improves insulin secretion in islets from type 2 diabetes patients and rescues beta-cell dysfunction caused by Hdac7 upregulation. Acta Diabetol. 55, 1231–1235 (2018).
Dayeh, T. et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 10, e1004160 (2014).
Dhawan, S. et al. DNA methylation directs functional maturation of pancreatic beta cells. J. Clin. Invest. 125, 2851–2860 (2015).
Hall, E. et al. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med. Genet. 14, 76 (2013).
Hall, E. et al. Glucolipotoxicity alters insulin secretion via epigenetic changes in human islets. Diabetes 68, 1965–1974 (2019).
Hall, E. et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 15, 522 (2014).
Malmgren, S. et al. Coordinate changes in histone modifications, mRNA levels, and metabolite profiles in clonal INS-1 832/13 beta-cells accompany functional adaptations to lipotoxicity. J. Biol. Chem. 288, 11973–11987 (2013).
Olsson, A. H. et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 10, e1004735 (2014).
Yang, B. T. et al. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol. Endocrinol. 26, 1203–1212 (2012).
Kameswaran, V. et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell. Metab. 19, 135–145 (2014).
Ling, C. et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 51, 615–622 (2008).
Volkmar, M. et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 31, 1405–1426 (2012).
Yang, B. T. et al. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 54, 360–367 (2011).
Kaneto, H. et al. PDX-1 functions as a master factor in the pancreas. Front. Biosci. 13, 6406–6420 (2008).
Lytrivi, M., Castell, A. L., Poitout, V. & Cnop, M. Recent insights into mechanisms of beta-cell lipo- and glucolipotoxicity in type 2 diabetes. J. Mol. Biol. 432, 1514–1534 (2020).
Hall, E. et al. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol. Cell Endocrinol. 472, 57–67 (2018).
Hall, E. et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 12, 103 (2014).
Carlsson, M., Wessman, Y., Almgren, P. & Groop, L. High levels of nonesterified fatty acids are associated with increased familial risk of cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 20, 1588–1594 (2000).
Tramunt, B. et al. Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia 63, 453–461 (2020).
Pasquali, L. et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46, 136–143 (2014).
Bhandare, R. et al. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 20, 428–433 (2010).
Parker, S. C. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA 110, 17921–17926 (2013).
Khetan, S. et al. Type 2 diabetes-associated genetic variants regulate chromatin accessibility in human islets. Diabetes 67, 2466–2477 (2018).
Varshney, A. et al. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl Acad. Sci. USA 114, 2301–2306 (2017).
Bysani, M. et al. ATAC-seq reveals alterations in open chromatin in pancreatic islets from subjects with type 2 diabetes. Sci. Rep. 9, 7785 (2019).
Khalid, M., Alkaabi, J., Khan, M. A. B. & Adem, A. Insulin signal transduction perturbations in insulin resistance. Int. J. Mol. Sci. 22, 8590 (2021).
Lillioja, S. et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N. Engl. J. Med. 329, 1988–1992 (1993).
James, D. E., Stockli, J. & Birnbaum, M. J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell Biol. 22, 751–771 (2021).
Petersen, M. C. & Shulman, G. I. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 98, 2133–2223 (2018).
Ryall, J. G. et al. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183 (2015).
Das, S. et al. ATP citrate lyase regulates myofiber differentiation and increases regeneration by altering histone acetylation. Cell Rep. 21, 3003–3011 (2017).
Davegardh, C. et al. VPS39-deficiency observed in type 2 diabetes impairs muscle stem cell differentiation via altered autophagy and epigenetics. Nat. Commun. 12, 2431 (2021).
Barres, R. et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 10, 189–198 (2009).
Brons, C. et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J. Clin. Endocrinol. Metab. 95, 3048–3056 (2010).
Bysani, M. et al. Epigenetic alterations in blood mirror age-associated DNA methylation and gene expression changes in human liver. Epigenomics 9, 105–122 (2017).
Davegårdh, C. et al. Abnormal epigenetic changes during differentiation of human skeletal muscle stem cells from obese subjects. BMC Med. 15, 39–39 (2017).
Dayeh, T. et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics 11, 482–488 (2016).
Gillberg, L. et al. Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia 59, 799–812 (2016).
Jacobsen, S. C. et al. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia 57, 1154–1158 (2014).
Ling, C. et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J. Clin. Invest. 117, 3427–3435 (2007).
Mudry, J. M. et al. Insulin and glucose alter death-associated protein kinase 3 (DAPK3) DNA methylation in human skeletal muscle. Diabetes 66, 651–662 (2017).
Nilsson, E. et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63, 2962–2976 (2014).
Nilsson, E. et al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocrinol. Metab. 100, E1491–E1501 (2015).
Nilsson, E. et al. Differential DNA methylation and expression of micrornas in adipose tissue from twin pairs discordant for type 2. Diabetes 70, 2402–2418 (2021).
Nitert, M. D. et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 61, 3322–3332 (2012).
Ribel-Madsen, R. et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE 7, e51302 (2012).
Ronn, T. et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 51, 1159–1168 (2008).
Ronn, T. et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum. Mol. Genet. 24, 3792–3813 (2015).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Taylor, D. L. et al. Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc. Natl Acad. Sci. USA 116, 10883–10888 (2019).
Chambers, J. C. et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 3, 526–534 (2015).
Peng, Y. et al. Phosphatase orphan 1 inhibits myoblast proliferation and promotes myogenic differentiation. FASEB J. 35, e21154 (2021).
Green, C. J., Pedersen, M., Pedersen, B. K. & Scheele, C. Elevated NF-kappaB activation is conserved in human myocytes cultured from obese type 2 diabetic patients and attenuated by AMP-activated protein kinase. Diabetes 60, 2810–2819 (2011).
Broholm, C. et al. Epigenetic programming of adipose-derived stem cells in low birthweight individuals. Diabetologia 59, 2664–2673 (2016).
Broholm, C. et al. Human adipogenesis is associated with genome-wide DNA methylation and gene-expression changes. Epigenomics 8, 1601–1617 (2016).
Keller, M. et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol. Metab. 6, 86–100 (2017).
You, D. et al. Dnmt3a is an epigenetic mediator of adipose insulin resistance. eLife 6, e30766 (2017).
Andersen, E. et al. Preadipocytes from obese humans with type 2 diabetes are epigenetically reprogrammed at genes controlling adipose tissue function. Int. J. Obes. 43, 306–318 (2019).
Hotamisligil, G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006).
Guenard, F. et al. Differential methylation in visceral adipose tissue of obese men discordant for metabolic disturbances. Physiol. Genomics 46, 216–222 (2014).
Castellano-Castillo, D. et al. Altered adipose tissue DNA methylation status in metabolic syndrome: relationships between global DNA methylation and specific methylation at adipogenic, lipid metabolism and inflammatory candidate genes and metabolic variables. J. Clin. Med. 8, 87 (2019).
Whincup, P. H. et al. Birth weight and risk of type 2 diabetes: a systematic review. JAMA 300, 2886–2897 (2008).
Hjort, L. et al. 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clin. Epigenetics 9, 40 (2017).
Arner, P. et al. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia 59, 2393–2405 (2016).
Rodriguez-Rodero, S. et al. Altered intragenic DNA methylation of HOOK2 gene in adipose tissue from individuals with obesity and type 2 diabetes. PLoS ONE 12, e0189153 (2017).
Crujeiras, A. B. et al. Genome-wide DNA methylation pattern in visceral adipose tissue differentiates insulin-resistant from insulin-sensitive obese subjects. Transl. Res. 178, 13–24 (2016).
Sievert, H. et al. Epigenetic downregulation of FASN in visceral adipose tissue of insulin resistant subjects. Exp. Clin. Endocrinol. Diabetes 129, 674–682 (2021).
Nadiger, N. et al. Protocol for a prospective, observational, deep phenotyping study on adipose epigenetic and lipidomic determinants of metabolic homoeostasis in South Asian Indians: the Indian Diabetes and Metabolic Health (InDiMeT) study. BMJ Open 11, e043644 (2021).
Sharma, N. K. et al. Integrative analysis of glucometabolic traits, adipose tissue DNA methylation, and gene expression identifies epigenetic regulatory mechanisms of insulin resistance and obesity in African Americans. Diabetes 69, 2779–2793 (2020).
Wewer Albrechtsen, N. J. et al. The liver-alpha-cell axis and type 2 diabetes. Endocr. Rev. 40, 1353–1366 (2019).
Abderrahmani, A. et al. Increased Hepatic PDGF-AA signaling mediates liver insulin resistance in obesity-associated type 2 diabetes. Diabetes 67, 1310–1321 (2018).
Krause, C. et al. Multi-layered epigenetic regulation of IRS2 expression in the liver of obese individuals with type 2 diabetes. Diabetologia 63, 2182–2193 (2020).
Kirchner, H. et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol. Metab. 5, 171–183 (2016).
Prokopenko, I. et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10, e1004235 (2014).
Zhang, N. et al. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 3, e120304 (2018).
Baumeier, C. et al. Hepatic DPP4 DNA methylation associates with fatty liver. Diabetes 66, 25–35 (2017).
Rusu, V. et al. Type 2 diabetes variants disrupt function of SLC16A11 through two distinct mechanisms. Cell 170, 199–212 (2017).
Lopez Rodriguez, M. et al. Identification and characterization of a FOXA2-regulated transcriptional enhancer at a type 2 diabetes intronic locus that controls GCKR expression in liver cells. Genome Med. 9, 63 (2017).
Knowler, W. C. et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 346, 393–403 (2002).
Ronn, T. et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 9, e1003572 (2013).
Lindholm, M. E. et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 9, 1557–1569 (2014).
Robinson, M. M. et al. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab. 25, 581–592 (2017).
Sailani, M. R. et al. Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci. Rep. 9, 3272 (2019).
Barres, R. et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411 (2012).
Fabre, O. et al. Exercise training alters the genomic response to acute exercise in human adipose tissue. Epigenomics 10, 1033–1050 (2018).
Bajpeyi, S. et al. Skeletal muscle PGC1alpha -1 nucleosome position and -260 nt DNA methylation determine exercise response and prevent ectopic lipid accumulation in men. Endocrinology 158, 2190–2199 (2017).
Ruple, B. A. et al. Resistance training rejuvenates the mitochondrial methylome in aged human skeletal muscle. FASEB J. 35, e21864 (2021).
Rowlands, D. S. et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in type 2 diabetic obesity. Physiol. Genomics 46, 747–765 (2014).
Stephens, N. A. et al. Exercise response variations in skeletal muscle PCr recovery rate and insulin sensitivity relate to muscle epigenomic profiles in individuals with type 2 diabetes. Diabetes Care 41, 2245–2254 (2018).
Denham, J., O’Brien, B. J., Marques, F. Z. & Charchar, F. J. Changes in the leukocyte methylome and its effect on cardiovascular-related genes after exercise. J. Appl. Physiol. 118, 475–488 (2015).
Fernandez-Sanles, A. et al. Physical activity and genome-wide DNA methylation: the REgistre GIroni del COR study. Med. Sci. Sports Exerc. 52, 589–597 (2020).
Hunter, D. J. et al. Impact of aerobic exercise and fatty acid supplementation on global and gene-specific DNA methylation. Epigenetics 14, 294–309 (2019).
Kankaanpaa, A. et al. Leisure-time and occupational physical activity associates differently with epigenetic aging. Med. Sci. Sports Exerc. 53, 487–495 (2021).
Kresovich, J. K. et al. Associations of body composition and physical activity level with multiple measures of epigenetic age acceleration. Am. J. Epidemiol. 190, 984–993 (2021).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019).
White, A. J. et al. Recreational and household physical activity at different time points and DNA global methylation. Eur. J. Cancer 49, 2199–2206 (2013).
Nakajima, K. et al. Exercise effects on methylation of ASC gene. Int. J. Sports Med. 31, 671–675 (2010).
Denham, J., O’Brien, B. J., Harvey, J. T. & Charchar, F. J. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics 7, 717–731 (2015).
Ingerslev, L. R. et al. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin. Epigenetics 10, 12 (2018).
Jonsson, J. et al. Lifestyle intervention in pregnant women with obesity impacts cord blood DNA methylation, which associates with body composition in the offspring. Diabetes 70, 854–866 (2021).
Antoun, E. et al. Maternal dysglycaemia, changes in the infant’s epigenome modified with a diet and physical activity intervention in pregnancy: secondary analysis of a randomised control trial. PLoS Med. 17, e1003229 (2020).
Jacobsen, S. C. et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia 55, 3341–3349 (2012).
Laker, R. C. et al. Transcriptomic and epigenetic responses to short-term nutrient-exercise stress in humans. Sci. Rep. 7, 15134 (2017).
Jorgensen, S. W. et al. Metabolic response to 36 hours of fasting in young men born small vs appropriate for gestational age. Diabetologia 58, 178–187 (2015).
Di Francesco, A., Di Germanio, C., Bernier, M. & de Cabo, R. A time to fast. Science 362, 770–775 (2018).
Gillberg, L. et al. Fasting unmasks differential fat and muscle transcriptional regulation of metabolic gene sets in low versus normal birth weight men. eBioMedicine 47, 341–351 (2019).
Fitzgerald, K. N. et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging 13, 9419–9432 (2021).
Quach, A. et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 9, 419–446 (2017).
Sun, D. et al. Genetic, epigenetic and transcriptional variations at NFATC2IP locus with weight loss in response to diet interventions: the POUNDS Lost trial. Diabetes Obes. Metab. 20, 2298–2303 (2018).
Rosqvist, F. et al. Overfeeding polyunsaturated and saturated fat causes distinct effects on liver and visceral fat accumulation in humans. Diabetes 63, 2356–2368 (2014).
Bjermo, H. et al. Effects of n-6 PUFAs compared with SFAs on liver fat, lipoproteins, and inflammation in abdominal obesity: a randomized controlled trial. Am. J. Clin. Nutr. 95, 1003–1012 (2012).
Perfilyev, A. et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am. J. Clin. Nutr. 105, 991–1000 (2017).
Arpon, A. et al. Impact of consuming extra-virgin olive oil or nuts within a mediterranean diet on DNA methylation in peripheral white blood cells within the PREDIMED-navarra randomized controlled trial: a role for dietary lipids. Nutrients 10, 15 (2017).
Corella, D., Coltell, O., Macian, F. & Ordovas, J. M. Advances in understanding the molecular basis of the mediterranean diet effect. Annu. Rev. Food Sci. Technol. 9, 227–249 (2018).
Ma, J. et al. Whole blood DNA methylation signatures of diet are associated with cardiovascular disease risk factors and all-cause mortality. Circ. Genom. Precis. Med. 13, e002766 (2020).
Phang, M. et al. Epigenetic aging in newborns: role of maternal diet. Am. J. Clin. Nutr. 111, 555–561 (2020).
Lai, C. Q. et al. Carbohydrate and fat intake associated with risk of metabolic diseases through epigenetics of CPT1A. Am. J. Clin. Nutr. 112, 1200–1211 (2020).
Voisin, S. et al. Meta-analysis of genome-wide DNA methylation and integrative omics of age in human skeletal muscle. J. Cachexia Sarcopenia Muscle 12, 1064–1078 (2021).
Zhou, J. et al. Elevated H3K27ac in aged skeletal muscle leads to increase in extracellular matrix and fibrogenic conversion of muscle satellite cells. Aging Cell 18, e12996 (2019).
Bigot, A. et al. Age-associated methylation suppresses SPRY1, leading to a failure of re-quiescence and loss of the reserve stem cell pool in elderly muscle. Cell Rep. 13, 1172–1182 (2015).
Blocquiaux, S. et al. Recurrent training rejuvenates and enhances transcriptome and methylome responses in young and older human muscle. JCSM Rapid Commun. 5, 10–32 (2022).
Horvath, S. et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci. USA 111, 15538–15543 (2014).
de Toro-Martin, J. et al. Body mass index is associated with epigenetic age acceleration in the visceral adipose tissue of subjects with severe obesity. Clin. Epigenetics 11, 172 (2019).
Loos, R. J. F. & Janssens, A. Predicting polygenic obesity using genetic information. Cell Metab. 25, 535–543 (2017).
Langenberg, C. & Lotta, L. A. Genomic insights into the causes of type 2 diabetes. Lancet 391, 2463–2474 (2018).
Volkov, P. et al. A genome-wide mQTL analysis in human adipose tissue identifies genetic variants associated with DNA methylation, gene expression and metabolic traits. PLoS ONE 11, e0157776 (2016).
Drong, A. W. et al. The presence of methylation quantitative trait loci indicates a direct genetic influence on the level of DNA methylation in adipose tissue. PLoS ONE 8, e55923 (2013).
Dayeh, T. A. et al. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia 56, 1036–1046 (2013).
Almen, M. S. et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics 99, 132–137 (2012).
Zhou, D. et al. Polymorphisms involving gain or loss of CpG sites are significantly enriched in trait-associated SNPs. Oncotarget 6, 39995–40004 (2015).
Shah, U. J. et al. Differential methylation of the type 2 diabetes susceptibility locus KCNQ1 is associated with insulin sensitivity and is predicted by CpG site specific genetic variation. Diabetes Res. Clin. Pract. 148, 189–199 (2019).
Wahl, S. et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541, 81–86 (2017).
Cardona, A. et al. Epigenome-wide association study of incident type 2 diabetes in a British population: EPIC-Norfolk study. Diabetes 68, 2315–2326 (2019).
Ouni, M. et al. Epigenetic changes in islets of Langerhans preceding the onset of diabetes. Diabetes 69, 2503–2517 (2020).
Agardh, E. et al. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 13, 182 (2015).
Juvinao-Quintero, D. L. et al. DNA methylation of blood cells is associated with prevalent type 2 diabetes in a meta-analysis of four European cohorts. Clin. Epigenetics 13, 40 (2021).
Galmozzi, A. et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 62, 732–742 (2013).
Gao, Z. et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517 (2009).
Lundh, M., Galbo, T., Poulsen, S. S. & Mandrup-Poulsen, T. Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes Obes. Metab. 17, 703–707 (2015).
Wagner, F. F. et al. An isochemogenic set of inhibitors to define the therapeutic potential of histone deacetylases in beta-cell protection. ACS Chem. Biol. 11, 363–374 (2016).
Kameswaran, V. et al. The dysregulation of the DLK1-MEG3 locus in islets from patients with type 2 diabetes is mimicked by targeted epimutation of its promoter with TALE-DNMT constructs. Diabetes 67, 1807–1815 (2018).
Liu, X. S. et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247 (2016).
Ou, K. et al. Targeted demethylation at the CDKN1C/p57 locus induces human beta cell replication. J. Clin. Invest. 129, 209–214 (2019).
Diabetes Prevention Program Research Group. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet 374, 1677–1686 (2009).
Soriano-Tarraga, C. et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum. Mol. Genet 25, 609–619 (2016).
Qie, R. et al. Association of ABCG1 gene methylation and its dynamic change status with incident type 2 diabetes mellitus: the Rural Chinese Cohort Study. J. Hum. Genet. 66, 347–357 (2021).
Kulkarni, H. et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum. Mol. Genet. 24, 5330–5344 (2015).
Kim, K. et al. DNA methylation grimage and incident diabetes: the coronary artery risk development in young adults (CARDIA) study. Diabetes 70, 1404–1413 (2021).
Feinberg, A. P. et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci. Transl. Med. 2, 49ra67 (2010).
Wang, X. et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 8, 87 (2010).
Dick, K. J. et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet 383, 1990–1998 (2014).
Agha, G. et al. Adiposity is associated with DNA methylation profile in adipose tissue. Int. J. Epidemiol. 44, 1277–1287 (2015).
Demerath, E. W. et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet. 24, 4464–4479 (2015).
Pan, H. et al. HIF3A association with adiposity: the story begins before birth. Epigenomics 7, 937–950 (2015).
Wang, S. et al. HIF3A DNA methylation is associated with childhood obesity and ALT. PLoS ONE 10, e0145944 (2015).
Richmond, R. C. et al. DNA methylation and BMI: investigating identified methylation sites at HIF3A in a causal framework. Diabetes 65, 1231–1244 (2016).
Mendelson, M. M. et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 14, e1002215 (2017).
Do, W. L. et al. Examining the association between adiposity and DNA methylation: a systematic review and meta-analysis. Obes. Rev. 22, e13319 (2021).
Godfrey, K. M. et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes 60, 1528–1534 (2011).
Relton, C. L. et al. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS ONE 7, e31821 (2012).
van Dijk, S. J. et al. DNA methylation in blood from neonatal screening cards and the association with BMI and insulin sensitivity in early childhood. Int. J. Obes. 42, 28–35 (2018).
Vehmeijer, F. O. L. et al. DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies. Genome Med. 12, 105 (2020).
Robinson, N. et al. Childhood DNA methylation as a marker of early life rapid weight gain and subsequent overweight. Clin. Epigenetics 13, 8 (2021).
Gutierrez-Repiso, C. et al. Epigenetic biomarkers of transition from metabolically healthy obesity to metabolically unhealthy obesity phenotype: a prospective study. Int. J. Mol. Sci. 22, 10417 (2021).
Esposito, K. et al. Inflammatory cytokine concentrations are acutely increased by hyperglycemia in humans: role of oxidative stress. Circulation 106, 2067–2072 (2002).
van Greevenbroek, M. M., Schalkwijk, C. G. & Stehouwer, C. D. Obesity-associated low-grade inflammation in type 2 diabetes mellitus: causes and consequences. Neth. J. Med. 71, 174–187 (2013).
Chung, W. K. et al. Precision medicine in diabetes: a Consensus Report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 63, 1671–1693 (2020).
Cook, M. N., Girman, C. J., Stein, P. P. & Alexander, C. M. Initial monotherapy with either metformin or sulphonylureas often fails to achieve or maintain current glycaemic goals in patients with type 2 diabetes in UK primary care. Diabet. Med. 24, 350–358 (2007).
Garcia-Calzon, S. et al. Epigenetic markers associated with metformin response and intolerance in drug-naive patients with type 2 diabetes. Sci. Transl. Med. 12, eaaz1803 (2020).
Lin, C. H., Lee, Y. S., Huang, Y. Y. & Tsai, C. N. Methylation status of vault RNA 2-1 promoter is a predictor of glycemic response to glucagon-like peptide-1 analog therapy in type 2 diabetes mellitus. BMJ Open Diabetes Res. Care 9, e001416 (2021).
Brown, A. et al. Dietary strategies for remission of type 2 diabetes: a narrative review. J. Hum. Nutr. Diet. 35, 165–178 (2022).
Cordero, P. et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J. Physiol. Biochem. 67, 463–470 (2011).
Stone, N. J. et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 63, 2889–2934 (2014).
Schrader, S., Perfilyev, A., Martinell, M., Garcia-Calzon, S. & Ling, C. Statin therapy is associated with epigenetic modifications in individuals with type 2 diabetes. Epigenomics 13, 919–925 (2021).
Ochoa-Rosales, C. et al. Epigenetic link between statin therapy and type 2 diabetes. Diabetes Care 43, 875–884 (2020).
Liu, Y. et al. Statin use associates with risk of type 2 diabetes via epigenetic patterns at ABCG1. Front. Genet. 11, 622 (2020).
Natarajan, R. Epigenetic mechanisms in diabetic vascular complications and metabolic memory: the 2020 Edwin Bierman Award Lecture. Diabetes 70, 328–337 (2021).
Qiu, C. et al. Cytosine methylation predicts renal function decline in American Indians. Kidney Int. 93, 1417–1431 (2018).
VanderJagt, T. A., Neugebauer, M. H., Morgan, M., Bowden, D. W. & Shah, V. O. Epigenetic profiles of pre-diabetes transitioning to type 2 diabetes and nephropathy. World J. Diabetes 6, 1113–1121 (2015).
Roshandel, D. et al. DNA methylation age calculators reveal association with diabetic neuropathy in type 1 diabetes. Clin. Epigenetics 12, 52 (2020).
Maghbooli, Z., Hossein-nezhad, A., Larijani, B., Amini, M. & Keshtkar, A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab. Res. Rev. 31, 183–189 (2015).
Allen, S. C. & Mamotte, C. D. S. Pleiotropic and adverse effects of statins — do epigenetics play a role? J. Pharmacol. Exp. Ther. 362, 319–326 (2017).
Bridgeman, S. C., Ellison, G. C., Melton, P. E., Newsholme, P. & Mamotte, C. D. S. Epigenetic effects of metformin: from molecular mechanisms to clinical implications. Diabetes Obes. Metab. 20, 1553–1562 (2018).
Garcia-Calzon, S. et al. Diabetes medication associates with DNA methylation of metformin transporter genes in the human liver. Clin. Epigenetics 9, 102 (2017).
Caton, P. W. et al. Metformin suppresses hepatic gluconeogenesis through induction of SIRT1 and GCN5. J. Endocrinol. 205, 97–106 (2010).
Khamis, A. et al. Histone deacetylase 9 promoter hypomethylation associated with adipocyte dysfunction is a statin-related metabolic effect. Clin. Epigenetics 12, 68 (2020).
Vallois, D. et al. Gluco-incretins regulate beta-cell glucose competence by epigenetic silencing of Fxyd3 expression. PLoS ONE 9, e103277 (2014).
Pinney, S. E., Jaeckle Santos, L. J., Han, Y., Stoffers, D. A. & Simmons, R. A. Exendin-4 increases histone acetylase activity and reverses epigenetic modifications that silence Pdx1 in the intrauterine growth retarded rat. Diabetologia 54, 2606–2614 (2011).
Cheng, Y. et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal. Transduct. Target. Ther. 4, 62 (2019).
Remsberg, J. R. et al. Deletion of histone deacetylase 3 in adult beta cells improves glucose tolerance via increased insulin secretion. Mol. Metab. 6, 30–37 (2017).
Hong, S. et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med. 23, 223–234 (2017).
Sun, Z. et al. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J. Biol. Chem. 286, 33301–33309 (2011).
Zhao, Z., Ukidve, A., Kim, J. & Mitragotri, S. Targeting strategies for tissue-specific drug delivery. Cell 181, 151–167 (2020).
Ling, C. & Ronn, T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 29, 1028–1044 (2019).
Acknowledgements
The authors’ research is supported by grants from the Swedish research council, Exodiab (2009-1039), Swedish Foundation for Strategic Research for IRC15-0067, the Diabetes Foundation, the European Foundation for the Study of Diabetes, the European Research Council (Paintbox), Region Skåne (ALF), the Novo Nordisk foundation, Påhlsson foundation, the EFSD/NNF and the Crafoord foundation.
Author information
Authors and Affiliations
Contributions
C.L., K.B. and T.R. contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Endocrinology thanks Henriette Kirchner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ling, C., Bacos, K. & Rönn, T. Epigenetics of type 2 diabetes mellitus and weight change — a tool for precision medicine?. Nat Rev Endocrinol 18, 433–448 (2022). https://doi.org/10.1038/s41574-022-00671-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41574-022-00671-w
This article is cited by
-
Advances in secondary prevention mechanisms of macrovascular complications in type 2 diabetes mellitus patients: a comprehensive review
European Journal of Medical Research (2024)
-
Highlights from SfE BES 2023
Nature Reviews Endocrinology (2024)
-
Association of low-carbohydrate diet scores and type 2 diabetes in Chinese rural adults: The Henan Rural Cohort Study
Endocrine (2024)
-
Epigenetic analyses in forensic medicine: future and challenges
International Journal of Legal Medicine (2024)
-
Effect of Zinc Supplementation on Lipid Profile and Body Composition in Patients with Type 2 Diabetes Mellitus: A GRADE-Assessed Systematic Review and Dose-Response Meta-analysis
Biological Trace Element Research (2024)