Abstract
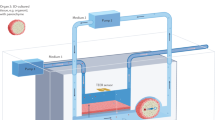
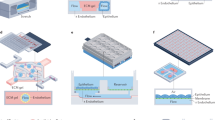
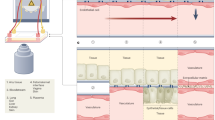
Organs-on-chips (OoCs), also known as microphysiological systems or ‘tissue chips’ (the terms are synonymous), have attracted substantial interest in recent years owing to their potential to be informative at multiple stages of the drug discovery and development process. These innovative devices could provide insights into normal human organ function and disease pathophysiology, as well as more accurately predict the safety and efficacy of investigational drugs in humans. Therefore, they are likely to become useful additions to traditional preclinical cell culture methods and in vivo animal studies in the near term, and in some cases replacements for them in the longer term. In the past decade, the OoC field has seen dramatic advances in the sophistication of biology and engineering, in the demonstration of physiological relevance and in the range of applications. These advances have also revealed new challenges and opportunities, and expertise from multiple biomedical and engineering fields will be needed to fully realize the promise of OoCs for fundamental and translational applications. This Review provides a snapshot of this fast-evolving technology, discusses current applications and caveats for their implementation, and offers suggestions for directions in the next decade.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203 (2010).
Scannell, J. W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191 (2012).
Seok, J. et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA 110, 3507–3512 (2013).
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40 (2014).
Waring, M. J. et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14, 475 (2015).
Sweeney, L. M., Shuler, M. L., Babish, J. G. & Ghanem, A. A cell culture analogue of rodent physiology: Application to naphthalene toxicology. Toxicol. In Vitro 9, 307–316 (1995).
Sin, A. et al. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 20, 338–345 (2004).
Huh, D. et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 (2010). Early OoC study recreating the alveolar–capillary interface of the human lung incorporating cyclic biomechanical stretch forces and showing replication of in vivo responses.
Rossi, G., Manfrin, A. & Lutolf, M. P. Progress and potential in organoid research. Nat. Rev. Genet. 19, 671–687 (2018).
Ingber, D. E. Reverse engineering human pathophysiology with organs-on-chips. Cell 164, 1105–1109 (2016).
Pamies, D. et al. A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. ALTEX https://doi.org/10.14573/altex.1609122 (2016).
Plummer, S. et al. A human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. 9, 1407 (2019).
Schwartz, M. P. et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl Acad. Sci. USA 112, 12516–12521 (2015).
Rothbauer, M., Rosser, J. M., Zirath, H. & Ertl, P. Tomorrow today: organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr. Opin. Biotechnol. 55, 81–86 (2019).
Kasendra, M. et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 8, 2871 (2018).
Ramme, A. P. et al. Towards an autologous iPSC-derived patient-on-a-chip. bioRxiv https://doi.org/10.1101/376970 (2018).
Vatine, G. D. et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 24, 995–1005.e1006 (2019).
Caliari, S. R. & Burdick, J. A. A practical guide to hydrogels for cell culture. Nat. Methods 13, 405 (2016).
Crapo, P. M., Tottey, S., Slivka, P. F. & Badylak, S. F. Effects of biologic scaffolds on human stem cells and implications for CNS tissue engineering. Tissue Eng. Part. A 20, 313–323 (2013).
Safaee, H. et al. Tethered jagged-1 synergizes with culture substrate stiffness to modulate notch-induced myogenic progenitor differentiation. Cell. Mol. Bioeng. 10, 501–513 (2017).
Trappmann, B. et al. Matrix degradability controls multicellularity of 3D cell migration. Nat. Commun. 8, 371 (2017).
Shin, D. S. et al. Synthesis of microgel sensors for spatial and temporal monitoring of protease activity. ACS Biomater. Sci. Eng. 4, 378–387 (2018).
Wikswo, J. P. et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans. Biomed. Eng. 60, 682–690 (2013).
Wikswo, J. P. et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 13, 3496–3511 (2013).
Johnson, B. P. et al. Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc. Natl Acad. Sci. USA 111, 18757 (2014).
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349 (2010).
Cyr, K. J., Avaldi, O. M. & Wikswo, J. P. Circadian hormone control in a human-on-a-chip: In vitro biology’s ignored component? Exp. Biol. Med. 242, 1714–1731 (2017).
Chang, S.-Y. et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight https://doi.org/10.1172/jci.insight.95978 (2017). Physically coupled liver and kidney OoCs used to uncover the mechanism of nephrotoxicity of aristolochic acid via bioactivation in the liver, showing utility of coupled OoCs for understanding toxic effects.
Phan, D. T. T. et al. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 17, 511–520 (2017).
Zhang, C., Zhao, Z., Abdul Rahim, N. A., van Noort, D. & Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip 9, 3185–3192 (2009).
Materne, E.-M. et al. The multi-organ chip - a microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. https://doi.org/10.3791/52526 (2015).
Maschmeyer, I. et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15, 2688–2699 (2015). Development of a multi-organ integrated OoC with pulsatile flow which reliably supports homeostasis over 28 days and allows ADME profiling and repeated dose drug testing.
Tsamandouras, N. et al. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS J. 19, 1499–1512 (2017).
Oleaga, C. et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 6, 20030 (2016). Development of a multi-organ pumpless OoC with common medium under serum-free conditions, maintaining tissues to 14 days and profiling accurate acute responses to therapeutic compounds.
Oleaga, C. et al. Human-on-a-chip systems: long-term electrical and mechanical function monitoring of a human-on-a-chip system (Adv. Funct. Mater. 8/2019). Adv. Funct. Mater. 29, 1970049 (2019).
Stone, H. A., Stroock, A. D. & Ajdari, A. Engineering flows in small devices: microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid Mech. 36, 381–411 (2004).
Lochovsky, C., Yasotharan, S. & Günther, A. Bubbles no more: in-plane trapping and removal of bubbles in microfluidic devices. Lab Chip 12, 595–601 (2012).
Kaarj, K. & Yoon, J.-Y. Methods of delivering mechanical stimuli to organ-on-a-chip. Micromachines https://doi.org/10.3390/mi10100700 (2019).
Kim, H. J., Huh, D., Hamilton, G. & Ingber, D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174 (2012).
Maoz, B. M. et al. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 17, 2294–2302 (2017).
Herland, A. et al. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS ONE 11, e0150360 (2016).
Musah, S. et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 1, 0069 (2017).
Agarwal, A., Goss, J. A., Cho, A., McCain, M. L. & Parker, K. K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 13, 3599–3608 (2013).
Nunes, S. S. et al. Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat. Methods 10, 781 (2013).
Lee-Montiel, F. T. et al. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp. Biol. Med. 242, 1617–1632 (2017).
Senutovitch, N. et al. Fluorescent protein biosensors applied to microphysiological systems. Exp. Biol. Med. 240, 795–808 (2015).
Zhang, Y. S. et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl Acad. Sci. USA 114, E2293 (2017). Advanced multi-organ platform for automated control and biosensing over multiple days, including pH, O2, temperature, protein biomarker presence and microscopes for imaging. Demonstrated cardiotoxicity and hepatotoxicity from chronic and acute drug dosing.
Toepke, M. W. & Beebe, D. J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 6, 1484–1486 (2006).
Markov, D. A., Lillie, E. M., Garbett, S. P. & McCawley, L. J. Variation in diffusion of gases through PDMS due to plasma surface treatment and storage conditions. Biomed. Microdevices 16, 91–96 (2014).
Tan, S. H., Nguyen, N.-T., Chua, Y. C. & Kang, T. G. Oxygen plasma treatment for reducing hydrophobicity of a sealed polydimethylsiloxane microchannel. Biomicrofluidics 4, 032204 (2010).
Chuah, Y. J. et al. Simple surface engineering of polydimethylsiloxane with polydopamine for stabilized mesenchymal stem cell adhesion and multipotency. Sci. Rep. 5, 18162 (2015).
van Meer, B. J. et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 482, 323–328 (2017).
Regehr, K. J. et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 9, 2132–2139 (2009).
Suntharalingam, G. et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 355, 1018–1028 (2006).
Kaur, R., Sidhu, P. & Singh, S. What failed BIA 10-2474 phase I clinical trial? Global speculations and recommendations for future phase I trials. J. Pharmacol. Pharmacother. 7, 120–126 (2016).
Fowler, S. et al. Microphysiological systems for ADME-related applications: current status and recommendations for system development and characterization. Lab Chip 20, 446–467 (2020).
Fabre, K. et al. Introduction to a manuscript series on the characterization and use of microphysiological systems (MPS) in pharmaceutical safety and ADME applications. Lab Chip 20, 1049–1057 (2020).
Rudmann, D. G. The emergence of microphysiological systems (organs-on-chips) as paradigm-changing tools for toxicologic pathology. Toxicol. Pathol. 47, 4–10 (2018).
Gerets, H. H. J. et al. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 28, 69–87 (2012).
Heslop, J. A. et al. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch. Toxicol. 91, 439–452 (2017).
Vernetti, L. A. et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 241, 101–114 (2016).
Li, X., George, S. M., Vernetti, L., Gough, A. H. & Taylor, D. L. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab Chip 18, 2614–2631 (2018).
Jang, K.-J. et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 11, eaax5516 (2019). First article to compare rat, dog and human microfluidic multicellular liver chips after exposure to hepatotoxic compounds. A fibrotic liver model showed species differences highlighting species-specific differences in drug metabolism and toxicity.
Mathur, A. et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 5, 8883 (2015).
Ahn, S. et al. Mussel-inspired 3D fiber scaffolds for heart-on-a-chip toxicity studies of engineered nanomaterials. Anal. Bioanal. Chem. 410, 6141–6154 (2018).
Marsano, A. et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 16, 599–610 (2016).
Lind, J. U. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 16, 303 (2016).
Capulli, A. K., MacQueen, L. A., O’Connor, B. B., Dauth, S. & Parker, K. K. Acute pergolide exposure stiffens engineered valve interstitial cell tissues and reduces contractility in vitro. Cardiovas. Pathol. 25, 316–324 (2016).
Goversen, B., van der Heyden, M. A. G., van Veen, T. A. B. & de Boer, T. P. The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: Special focus on IK1. Pharmacol. Ther. 183, 127–136 (2018).
Sheehy, S. P. et al. Toward improved myocardial maturity in an organ-on-chip platform with immature cardiac myocytes. Exp. Biol. Med. 242, 1643–1656 (2017).
Ronaldson-Bouchard, K. et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 (2018). OoC used to show enhanced maturation of stem cell-derived cardiac tissues in three dimensions when subjected to electrical stimulation protocols, addressing a key challenge in cardiac stem cell differentiation.
Zhao, Y. et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913–927.e918 (2019).
Mills, R. J. et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl Acad. Sci. USA 114, E8372 (2017).
Giacomelli, E. et al. Human-iPSC-derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 26, 862–879.e811 (2020).
Jang, K.-J. et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 5, 1119–1129 (2013).
Kim, S. et al. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 8, 015021 (2016).
Weber, E. J. et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 90, 627–637 (2016).
Weber, E. J. et al. Human kidney on a chip assessment of polymyxin antibiotic nephrotoxicity. JCI Insight https://doi.org/10.1172/jci.insight.123673 (2018).
Kim, H. J. & Ingber, D. E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 5, 1130–1140 (2013).
Workman, M. J. et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49 (2016).
Williamson, I. A. et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol. 6, 301–319 (2018).
Shah, P. et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 7, 11535 (2016).
Workman, M. J. et al. Enhanced utilization of induced pluripotent stem cell–derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 5, 669–677.e662 (2018).
Tovaglieri, A. et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 7, 43 (2019).
Jalili-Firoozinezhad, S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019).
Benam, K. H. et al. in 3D Cell Culture: Methods and Protocols (ed. Koledova, Z.) 345–365 (Springer, 2017).
Benam, K. H. et al. Matched-comparative modeling of normal and diseased human airway responses using a microengineered breathing lung chip. Cell Syst. 3, 456–466.e454 (2016).
Blundell, C. et al. Placental drug transport-on-a-chip: a microengineered in vitro model of transporter-mediated drug efflux in the human placental barrier. Adv. Healthc. Mater. 7, 1700786 (2018).
Yin, F. et al. A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol. In Vitro 54, 105–113 (2019).
Sobrino, A. et al. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 6, 31589 (2016).
Barrile, R. et al. Organ-on-chip recapitulates thrombosis induced by an anti-CD154 monoclonal antibody: translational potential of advanced microengineered systems. Clin. Pharmacol. Ther. 104, 1240–1248 (2018).
Cook, D. et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 13, 419–431 (2014).
Horton, R. E. et al. Angiotensin II induced cardiac dysfunction on a chip. PLoS ONE 11, e0146415 (2016).
Hinson, J. T. et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986 (2015).
Nesmith, A. P., Agarwal, A., McCain, M. L. & Parker, K. K. Human airway musculature on a chip: an in vitro model of allergic asthmatic bronchoconstriction and bronchodilation. Lab Chip 14, 3925–3936 (2014).
van der Meer, A. D., Orlova, V. V., ten Dijke, P., van den Berg, A. & Mummery, C. L. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip 13, 3562–3568 (2013).
Cruz, N. M. et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 16, 1112 (2017).
Faal, T. et al. Induction of mesoderm and neural crest-derived pericytes from human pluripotent stem cells to study blood-brain barrier interactions. Stem Cell Rep. 12, 451–460 (2019).
Rooney, G. E. et al. Human iPS cell-derived neurons uncover the impact of increased ras signaling in Costello syndrome. J. Neurosci. 36, 142 (2016).
Atchison, L., Zhang, H., Cao, K. & Truskey, G. A. A tissue engineered blood vessel model of Hutchinson-Gilford progeria syndrome using human iPSC-derived smooth muscle cells. Sci. Rep. 7, 8168 (2017).
Wang, G. et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014). Rare paediatric disease modelled on an OoC and mechanism of disease uncovered using gene editing techniques.
Ben Jehuda, R., Shemer, Y. & Binah, O. Genome editing in induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rev. Rep. 14, 323–336 (2018).
Nguyen, D.-H. T. et al. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl Acad. Sci. USA 110, 6712–6717 (2013).
Montanez-Sauri, S. I., Sung, K. E., Berthier, E. & Beebe, D. J. Enabling screening in 3D microenvironments: probing matrix and stromal effects on the morphology and proliferation of T47D breast carcinoma cells. Integr. Biol. 5, 631–640 (2013).
Zervantonakis, I. K. et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl Acad. Sci. USA 109, 13515 (2012).
Jeon, J. S. et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl Acad. Sci. USA 112, 214–219 (2015).
Clark, A. M. et al. A model of dormant-emergent metastatic breast cancer progression enabling exploration of biomarker signatures. Mol. Cell. Proteomics 17, 619 (2018).
Shirure, V. S. et al. Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab Chip 18, 3687–3702 (2018).
Regier, M. C. et al. Transitions from mono- to co- to tri-culture uniquely affect gene expression in breast cancer, stromal, and immune compartments. Biomed. Microdevices 18, 70 (2016).
Marturano-Kruik, A. et al. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl Acad. Sci. USA 115, 1256 (2018).
Kim, S., Lee, H., Chung, M. & Jeon, N. L. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 13, 1489–1500 (2013).
Miller, C. P., Tsuchida, C., Zheng, Y., Himmelfarb, J. & Akilesh, S. A 3D human renal cell carcinoma-on-a-chip for the study of tumor angiogenesis. Neoplasia 20, 610–620 (2018).
Hassell, B. A. et al. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 21, 508–516 (2017).
Lee, J.-H. et al. Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance. J. Exp. Clin. Cancer Res. 37, 4 (2018).
Jeong, S.-Y., Lee, J.-H., Shin, Y., Chung, S. & Kuh, H.-J. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS ONE 11, e0159013 (2016).
Rizvi, I. et al. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc. Natl Acad. Sci. USA 110, E1974 (2013).
Li, R. et al. Macrophage-secreted TNFα and TGFβ1 influence migration speed and persistence of cancer cells in 3D tissue culture via independent pathways. Cancer Res. 77, 279 (2017).
Wang, N. et al. 3D microfluidic in vitro model and bioinformatics integration to study the effects of Spatholobi Caulis tannin in cervical cancer. Sci. Rep. 8, 12285 (2018).
Low, L. A. & Tagle, D. A. Tissue chips to aid drug development and modeling for rare diseases. Expert Opin. Orphan Drugs 4, 1113–1121 (2016).
Shik Mun, K. et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 10, 3124 (2019).
Shahjalal, H. M., Abdal Dayem, A., Lim, K. M., Jeon, T.-i & Cho, S.-G. Generation of pancreatic β cells for treatment of diabetes: advances and challenges. Stem Cell Res. Ther. 9, 355 (2018).
Takebe, T., Zhang, B. & Radisic, M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell 21, 297–300 (2017).
Park, S. E., Georgescu, A. & Huh, D. Organoids-on-a-chip. Science 364, 960 (2019).
Takebe, T. et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 16, 556–565 (2015).
Zhang, Y. S. et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110, 45–59 (2016).
Park, D., Lee, J., Chung, J. J., Jung, Y. & Kim, S. H. Integrating organs-on-chips: multiplexing, scaling, vascularization, and innervation. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2019.06.006 (2019).
Edington, C. D. et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 8, 4530 (2018). Multi-organ microfluidic ‘physiome-on-a-chip’ platform modelling up to 10 organs for 4 weeks with pharmacokinetic analysis of diclofenac metabolism, noting general design and operational principles for multi-organ platforms.
Novak, R. et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 4, 407–420 (2020). Multi-organ linked system for up to 10 OoCs for 3 weeks in automated culture and perfusion machine capable of medium addition, fluidic linking, sample collection and in situ microscopy.
Shim, M. K. et al. Carrier-free nanoparticles of cathepsin B-cleavable peptide-conjugated doxorubicin prodrug for cancer targeting therapy. J. Control. Release 294, 376–389 (2019).
Al-Malahmeh, A. J. et al. Physiologically based kinetic modeling of the bioactivation of myristicin. Arch. Toxicol. 91, 713–734 (2017).
Schurdak, M. E. et al. in Phenotypic Screening: Methods and Protocols (ed. Wagner B.) 207–222 (Springer, 2018).
Oliver, C. R. et al. A platform for artificial intelligence based identification of the extravasation potential of cancer cells into the brain metastatic niche. Lab Chip https://doi.org/10.1039/C8LC01387J (2019).
Satoh, T. et al. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip 18, 115–125 (2018).
Boos, J. A., Misun, P. M., Michlmayr, A., Hierlemann, A. & Frey, O. Microfluidic multitissue platform for advanced embryotoxicity testing in vitro. Adv. Sci. 6, 1900294–1900294 (2019).
Vunjak-Novakovic, G., Bhatia, S., Chen, C. & Hirschi, K. HeLiVa platform: integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res. Ther. 4, 1–6 (2013).
Vernetti, L. et al. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci. Rep. 7, 42296 (2017).
Brown, J. A. et al. Recreating blood-brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics 9, 054124 (2015).
Brown, J. A. et al. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J. Neuroinflammation 13, 306 (2016).
Maoz, B. M. et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 36, 865 (2018).
Chen, W. L. K. et al. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol. Bioeng. 114, 2648–2659 (2017).
Esch, M. B., Ueno, H., Applegate, D. R. & Shuler, M. L. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 16, 2719–2729 (2016).
Loskill, P., Marcus, S. G., Mathur, A., Reese, W. M. & Healy, K. E. μOrgano: a Lego®-like plug & play system for modular multi-organ-chips. PLoS ONE 10, e0139587 (2015).
Xiao, S. et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat. Commun. 8, 14584 (2017). Human female reproductive system and cycle recreated on a five-organ platform with inclusion of endocrine signalling to mimic hormonal markers of pregnancy as a tool for female reproductive toxicity assessment.
Skardal, A., Shupe, T. & Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 21, 1399–1411 (2016).
Skardal, A. et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 7, 8837 (2017).
Oleaga, C. et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 182, 176–190 (2018).
Sakolish, C. et al. Technology transfer of the microphysiological systems: a case study of the human proximal tubule tissue chip. Sci. Rep. 8, 14882–14882 (2018).
Roberts, R. A. et al. Reducing attrition in drug development: smart loading preclinical safety assessment. Drug Discov. Today 19, 341–347 (2014).
Livingston, C. A., Fabre, K. M. & Tagle, D. A. Facilitating the commercialization and use of organ platforms generated by the microphysiological systems (Tissue Chip) program through public–private partnerships. Comput. Struct. Biotechnol. J. 14, 207–210 (2016).
Schurdak, M. et al. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. Lab Chip 20, 1472–1492 (2020).
Lee, J. et al. Recent advances in genome editing of stem cells for drug discovery and therapeutic application. Pharmacol. Ther. https://doi.org/10.1016/j.pharmthera.2020.107501 (2020).
Franzen, N. et al. Impact of organ-on-a-chip technology on pharmaceutical R&D costs. Drug Discov. Today 24, 1720–1724 (2019).
Sances, S. et al. Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep. 10, 1222–1236 (2018). Increased calcium transients and mature gene expression seen in spinal motor neurons and brain microvascular endothelial cells derived from iPS cells when cultured on a 3D OoC versus a 96-well plate.
Mulholland, T. et al. Drug screening of biopsy-derived spheroids using a self-generated microfluidic concentration gradient. Sci. Rep. 8, 14672 (2018).
Schutgens, F. et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313 (2019).
Ronaldson-Bouchard, K. & Vunjak-Novakovic, G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 22, 310–324 (2018).
McAleer, C. W. et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 11, eaav1386 (2019).
Wagner, J. A. et al. Application of a dynamic map for learning, communicating, navigating, and improving therapeutic development. Clin. Transl. Sci. 11, 166–174 (2017).
Wagner, J. A. et al. Drug Discovery, Development and Deployment Map (4DM): Small Molecules. National Center for Advancing Translational Sciences https://ncats.nih.gov/translation/maps (NIH).
Peterson, N. C., Mahalingaiah, P. K., Fullerton, A. & Di Piazza, M. Application of microphysiological systems in biopharmaceutical research and development. Lab Chip 20, 697–708 (2020).
Hardwick, R. N. et al. Drug-induced skin toxicity: gaps in preclinical testing cascade as opportunities for complex in vitro models and assays. Lab Chip 20, 199–214 (2020).
Ainslie, G. R. et al. Microphysiological lung models to evaluate the safety of new pharmaceutical modalities: a biopharmaceutical perspective. Lab Chip 19, 3152–3161 (2019).
Peters, M. F. et al. Developing in vitro assays to transform gastrointestinal safety assessment: potential for microphysiological systems. Lab Chip 20, 1177–1190 (2020).
Phillips, J. A. et al. A pharmaceutical industry perspective on microphysiological kidney systems for evaluation of safety for new therapies. Lab Chip 20, 468–476 (2020).
Baudy, A. R. et al. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip 20, 215–225 (2020).
Yeung, C. K. et al. Tissue chips in space — challenges and opportunities. Clin. Transl. Sci. https://doi.org/10.1111/cts.12689 (2019).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010).
Author information
Authors and Affiliations
Contributions
L.A.L. wrote and edited the manuscript and created the figures. C.M., B.R.B., D.A.T. and C.P.A. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CiPA: http://cipaproject.org/
Defense Advanced Research Project Agency (DARPA) funded linked 10-organ system: https://www.darpa.mil/program/microphysiological-systems
EUROoCS: https://euroocs.eu/
European Union Reference Laboratory for Alternatives to Animal Testing: https://ec.europa.eu/jrc/en/eurl/ecvam
Good manufacturing practice guidelines: https://www.ecfr.gov/cgi-bin/text-idx?SID=cb7c830642b365274d824a432e118e77&mc=true&node=pt21.8.820&rgn=div5
Human Organ and Disease Model Technologies (hDMT): https://www.hdmt.technology/
IQ Consortium: https://iqconsortium.org/
National Center for Advancing Translational Sciences (NCATS) Tissue Chips in Space: https://ncats.nih.gov/tissuechip/projects/space
NIH Heal Initiative: https://heal.nih.gov
ORCHID: https://h2020-orchid.eu/
Organisation for Economic Co-operation and Development “Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment”: https://ntp.niehs.nih.gov/iccvam/suppdocs/feddocs/oecd/oecd-gd34.pdf
Organs on chips — 2017 market overview analysis by Yole Développement: http://www.yole.fr/OrgansOnChips_Market.aspx#.XIP6dVNKiV4
The Center for Advancement of Science in Space (CASIS): https://www.iss-casis.org/
The Gartner hype cycle for emerging technologies 2018: https://www.gartner.com/smarterwithgartner/5-trends-emerge-in-gartner-hype-cycle-for-emerging-technologies-2018/
Tissue Chip Drug Screening: https://ncats.nih.gov/tissuechip
Tissue Chip Testing Centers: https://ncats.nih.gov/tissuechip/projects/Centres
US National Institutes of Health (NIH), FDA and DARPA funded development of tissue chips to advance regulatory sciences: https://www.nih.gov/news-events/news-releases/nih-fda-announce-collaborative-initiative-fast-track-innovations-public
Glossary
- Extracellular matrix
-
(ECM). Supporting network of macromolecules providing structural and biochemical support to surrounding cells. Promotes cell adhesion and cell–cell communication and produces biochemical cues for tissue growth and maintenance. The ECM is tissue specific and in animal tissues consists of fibrous elements (collagen and elastin), and links proteins (laminin and fibronectin) and other molecules.
- Hydrogels
-
Highly absorbent and hydrophilic biocompatible 3D polymer networks used to contain cells or drugs for tissue engineering applications. Can consist of natural (collagen, gelatin and agarose) or synthetic components and respond to environmental conditions such as pH. May have both liquid and solid properties. Other uses include wound dressings and contact lenses.
- Multi-electrode arrays
-
Arrays of tens to thousands of tightly spaced microelectrical sensors designed to record from single cells to networks of cells on submillisecond timescales. Can also be used to stimulate cells with precise spatial and temporal characteristics. Used in electrically excitable tissues such as cardiac, muscle and neural tissues.
- Pharmacokinetic/pharmacodynamic modelling
-
Integration of pharmacokinetics (movement of drugs through the body) and pharmacodynamics (the body’s biological response to drugs) into a mathematical model describing dose–concentration–response relationships. Can be used to predict effect and efficacy of drug dosing over time.
- Physiologically based pharmacokinetic modelling
-
Mathematical modelling of body compartments (predefined organs or tissues) combined with known parameters of concentrations, quantities and transport between compartments used to predict absorption, distribution, metabolism and excretion of synthetic or natural chemical substances within the body.
- Investigational New Drug
-
(IND). An application submitted to the US Food and Drug Administration to administer a novel drug to humans. The first step in the drug review process, which includes information on animal studies, manufacturing protocols and clinical and personnel protocols. Data gathered become part of the New Drug Application.
- New Drug Application
-
(NDA). An application submitted to the US Food and Drug Administration requesting permission to sell and market a drug in the USA. Information submitted includes data from the Investigational New Drug and is reviewed for safety and efficacy, benefit versus risks, appropriate labelling information, and manufacturing and processing methods.
Rights and permissions
About this article
Cite this article
Low, L.A., Mummery, C., Berridge, B.R. et al. Organs-on-chips: into the next decade. Nat Rev Drug Discov 20, 345–361 (2021). https://doi.org/10.1038/s41573-020-0079-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-020-0079-3
This article is cited by
-
Microfluidic high-throughput 3D cell culture
Nature Reviews Bioengineering (2024)
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Integration of multiple flexible electrodes for real-time detection of barrier formation with spatial resolution in a gut-on-chip system
Microsystems & Nanoengineering (2024)
-
Replacing Animal Testing with Stem Cell-Organoids : Advantages and Limitations
Stem Cell Reviews and Reports (2024)
-
Passive-Flow-Based MPS: Emerging Physiological Flow-Mimetic Platforms for Studying Effects of Flow on Single Tissues and Inter-tissue Interactions
BioChip Journal (2024)