Abstract
The development of immunotherapies over the past decade has resulted in a paradigm shift in the treatment of cancer. However, the majority of patients do not benefit from immunotherapy, presumably owing to insufficient reprogramming of the immunosuppressive tumour microenvironment (TME) and thus limited reinvigoration of antitumour immunity. Various metabolic machineries and nutrient-sensing mechanisms orchestrate the behaviour of immune cells in response to nutrient availability in the TME. Notably, tumour-infiltrating immune cells typically experience metabolic stress as a result of the dysregulated metabolic activity of tumour cells, leading to impaired antitumour immune responses. Moreover, the immune checkpoints that are often exploited by tumour cells to evade immunosurveillance have emerging roles in modulating the metabolic and functional activity of T cells. Thus, repurposing of drugs targeting cancer metabolism might synergistically enhance immunotherapy via metabolic reprogramming of the TME. In addition, interventions targeting the metabolic circuits that impede antitumour immunity have been developed, with several clinical trials underway. Herein, we discuss how these metabolic circuits regulate antitumour immunity and the possible approaches to targeting these pathways in the context of anticancer immunotherapy. We also describe hypothetical combination treatments that could be used to better unleash the potential of adoptive cell therapies by enhancing T cell metabolism.
Key points
-
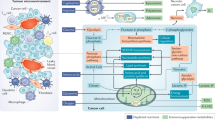
Conditions in the tumour microenvironment (TME) can impose metabolic stress on infiltrating immune cells, which can result in local immunosuppression and tumour immune evasion.
-
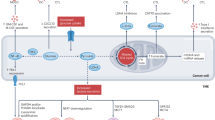
Immune checkpoints mediated by either co-activatory or inhibitory receptors modulate T cell activation and function, in part, by influencing metabolic reprogramming and mitochondrial fitness in these cells.
-
Agents targeting the interacting and competing metabolic pathways that are active in the TME might synergize with immune-checkpoint inhibitors by alleviating metabolic stress in tumour-infiltrating lymphocytes (TILs).
-
Thus, interventions targeting aberrant metabolic properties of tumour cells might reprogramme the immune state of the TME, in particular, via direct and indirect effects on myeloid cells.
-
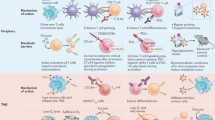
Modulation of the metabolic programme of T cells during ex vivo TIL expansion or the manufacturing of chimeric antigen receptor (CAR) T cells is a promising strategy to improve efficacy of adoptive T cell-based immunotherapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Boroughs, L. K. & DeBerardinis, R. J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351 (2015).
Lorendeau, D., Christen, S., Rinaldi, G. & Fendt, S.-M. Metabolic control of signaling pathways and metabolic auto-regulation. Biol. Cell 107, 251–272 (2015).
Rinaldi, G., Rossi, M. & Fendt, S.-M. Metabolic interactions in cancer: cellular metabolism at the interface between the microenvironment, the cancer cell phenotype and the epigenetic landscape. Wiley Interdiscip. Rev. Syst. Biol. Med. 10, e1397 (2018).
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Geeraerts, X., Bolli, E., Fendt, S.-M. & Van Ginderachter, J. A. Macrophage metabolism as therapeutic target for cancer, atherosclerosis, and obesity. Front. Immunol. 8, 289 (2017).
Sullivan, L. B., Gui, D. Y. & Heiden, M. G. V. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat. Rev. Cancer 16, 680 (2016).
Sciacovelli, M. & Frezza, C. Oncometabolites: unconventional triggers of oncogenic signalling cascades. Free Radic. Biol. Med. 100, 175–181 (2016).
Haas, R. et al. Intermediates of metabolism: from bystanders to signalling molecules. Trends Biochem. Sci. 41, 460–471 (2016).
Murphy, M. P. & O’Neill, L. A. J. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell 174, 780–784 (2018).
Shimizu, T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49, 123–150 (2009).
Dennis, E. A. & Norris, P. C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 15, 511–523 (2015).
Bi, J., Wu, S., Zhang, W. & Mischel, P. S. Targeting cancer’s metabolic co-dependencies: a landscape shaped by genotype and tissue context. Biochim. Biophys. Acta 1870, 76–87 (2018).
Elia, I., Schmieder, R., Christen, S. & Fendt, S.-M. Organ-specific cancer metabolism and its potential for therapy. Handb. Exp. Pharmacol. 233, 321–353 (2016).
Elia, I. & Fendt, S.-M. In vivo cancer metabolism is defined by the nutrient microenvironment. Transl Cancer Res. 5, S1284–S1287 (2016).
Muir, A., Danai, L. V. & Vander Heiden, M. G. Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis. Model. Mech. 11, dmm035758 (2018).
Elia, I., Doglioni, G. & Fendt, S.-M. Metabolic hallmarks of metastasis formation. Trends Cell Biol. 28, 673–684 (2018).
Lunt, S. Y. & Fendt, S.-M. Metabolism — a cornerstone of cancer initiation, progression, immune evasion and treatment response. Curr. Opin. Syst. Biol. 8, 67–72 (2018).
Heiden, M. G. V. & DeBerardinis, R. J. Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017).
Teoh, S. T. & Lunt, S. Y. Metabolism in cancer metastasis: bioenergetics, biosynthesis, and beyond. Wiley Interdiscip. Rev. Syst. Biol. Med. 10, e1406 (2018).
Warburg, O. On the origin of cancer cells. Science 123, 309 (1956).
Halestrap, A. P. The monocarboxylate transporter family — structure and functional characterization. IUBMB Life 64, 1–9 (2012).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015).
Li, W. et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 28, 87–103 (2018).
Dietl, K. et al. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. 184, 1200–1209 (2010).
Gottfried, E. et al. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107, 2013–2021 (2006).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014).
Husain, Z., Huang, Y., Seth, P. & Sukhatme, V. P. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells. J. Immunol. 191, 1486–1495 (2013).
Shime, H. et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J. Immunol. 180, 7175–7183 (2008).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115 (2017).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371 (2017).
Cascone, T. et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987 (2018).
Brand, A. et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 24, 657–671 (2016).
Liu, R. et al. Overall survival of cancer patients with serum lactate dehydrogenase greater than 1000 IU/L. Tumor Biol. 37, 14083–14088 (2016).
Walenta, S. et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 60, 916–921 (2000).
Christofk, H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008).
Elf, S. E. & Chen, J. Targeting glucose metabolism in patients with cancer. Cancer 120, 774–780 (2014).
Kung, C. et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem. Biol. 19, 1187–1198 (2012).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 is required for the expression of the immune checkpoint PD-L1 in immune cells and tumors. Front. Immunol. 8, 1300 (2017).
Li, F.-L. et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat. Commun. 9, 508 (2018).
Li, H.-M. et al. Blockage of glycolysis by targeting PFKFB3 suppresses tumor growth and metastasis in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 36, 7 (2017).
Chesney, J. A., Telang, S., Yaddanapudi, K. & Grewal, J. S. Targeting 6-phosphofructo-2-kinase (PFKFB3) as an immunotherapeutic strategy. J. Clin. Oncol. 34, e14548 (2016).
Yang, M. et al. HIF-dependent induction of adenosine receptor A2b skews human dendritic cells to a Th2-stimulating phenotype under hypoxia. Immunol. Cell Biol. 88, 165–171 (2010).
Deck, L. M. et al. Selective inhibitors of human lactate dehydrogenases and lactate dehydrogenase from the malarial parasite plasmodium falciparum. J. Med. Chem. 41, 3879–3887 (1998).
Weide, B. et al. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin. Cancer Res. 22, 5487–5496 (2016).
Murray, C. M. et al. Monocarboxylate transporter MCT1 is a target for immunosuppression. Nat. Chem. Biol. 1, 371–376 (2005).
Pilon-Thomas, S. et al. Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Res. 76, 1381–1390 (2016).
Pérez-Escuredo, J. et al. Lactate promotes glutamine uptake and metabolism in oxidative cancer cells. Cell Cycle 15, 72–83 (2016).
Ko, Y.-H. et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells. Cancer Biol. Ther. 12, 1085–1097 (2011).
Xiang, Y. et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306 (2015).
Wang, J. B. et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 (2010).
Gross, M. I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 (2014).
DeLaBarre, B., Hurov, J., Cianchetta, G., Murray, S. & Dang, L. Action at a distance: allostery and the development of drugs to target cancer cell metabolism. Chem. Biol. 21, 1143–1161 (2014).
Carr, E. L. et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 (2010).
Newsholme, P., Gordon, S. & Newsholme, E. A. Rates of utilization and fates of glucose, glutamine, pyruvate, fatty acids and ketone bodies by mouse macrophages. Biochem. J. 242, 631–636 (1987).
Liu, P.-S. et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985 (2017).
Nabe, S. et al. Reinforce the antitumor activity of CD8+ T cells via glutamine restriction. Cancer Sci. 109, 3737–3750 (2018).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018).
Tannir, N. M. et al. CANTATA: a randomized phase 2 study of CB-839 in combination with cabozantinib versus placebo with cabozantinib in patients with advanced/metastatic renal cell carcinoma. J. Clin. Oncol. 36, TPS4601 (2018).
Klysz, D. et al. Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal. 8, ra97 (2015).
Shanker, A., de Aquino, M. T. P., Hodo, T. & Uzhachenko, R. Glutamate receptors provide costimulatory signals to improve T cell immune response. J. Immunol. 200 (Suppl), 47.24 (2018).
Poulopoulou, C. et al. Modulation of voltage-gated potassium channels in human T lymphocytes by extracellular glutamate. Mol. Pharmacol. 67, 856–867 (2005).
Grohmann, U. et al. Amino-acid sensing and degrading pathways in immune regulation. Cytokine Growth Factor Rev. 35, 37–45 (2017).
Speiser, D. E., Ho, P. C. & Verdeil, G. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 16, 599–611 (2016).
He, X., Lin, H., Yuan, L. & Li, B. Combination therapy with L-arginine and alpha-PD-L1 antibody boosts immune response against osteosarcoma in immunocompetent mice. Cancer Biol. Ther. 18, 94–100 (2017).
Geiger, R. et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842 (2016).
Steggerda, S. M. et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J. Immunother. Cancer 5, 101 (2017).
Qiu, F. et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci. Signal. 7, ra31 (2014).
Kelly, M. P. et al. Arginine deiminase PEG20 inhibits growth of small cell lung cancers lacking expression of argininosuccinate synthetase. Br. J. Cancer 106, 324–332 (2011).
Tsai, H.-J. et al. A phase II study of arginine deiminase (ADI-PEG20) in relapsed/refractory or poor-risk acute myeloid leukemia patients. Sci. Rep. 7, 11253 (2017).
Brin, E. et al. PEGylated arginine deiminase can modulate tumor immune microenvironment by affecting immune checkpoint expression, decreasing regulatory T cell accumulation and inducing tumor T cell infiltration. Oncotarget 8, 58948–58963 (2017).
Liu, H. et al. Increased expression of IDO associates with poor postoperative clinical outcome of patients with gastric adenocarcinoma. Sci. Rep. 6, 21319 (2016).
Mbongue, J. C. et al. The role of indoleamine 2, 3-dioxygenase in immune suppression and autoimmunity. Vaccines 3, 703–729 (2015).
Munn, D. H. et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642 (2005).
Li, R. et al. IDO inhibits T-cell function through suppressing Vav1 expression and activation. Cancer Biol. Ther. 8, 1402–1408 (2009).
Cronin, S. J. F. et al. The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature 563, 564–568 (2018).
Minhas, P. S. et al. Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat. Immunol. 20, 50–63 (2019).
Zheng, X. et al. Silencing IDO in dendritic cells: a novel approach to enhance cancer immunotherapy in a murine breast cancer model. Int. J. Cancer 132, 967–977 (2013).
Yen, M.-C. et al. A novel cancer therapy by skin delivery of indoleamine 2,3-dioxygenase siRNA. Clin. Cancer Res. 15, 641–649 (2009).
Huang, T.-T. et al. Skin delivery of short hairpin RNA of indoleamine 2,3 dioxygenase induces antitumor immunity against orthotopic and metastatic liver cancer. Cancer Sci. 102, 2214–2220 (2011).
Soliman, H., Mediavilla-Varela, M. & Antonia, S. Indoleamine 2,3-dioxygenase: is it an immune suppressor? Cancer J. 16, 354–359 (2010).
Zakharia, Y. et al. Results of phase 1b trial of the indoleamine 2,3-dioxygenase (IDO) pathway inhibitor indoximod plus ipilimumab for the treatment of unresectable stage III or IV melanoma. Eur. J. Cancer 51, S108 (2015).
Yue, E. W. et al. INCB24360 (epacadostat), a highly potent and selective indoleamine-2,3-dioxygenase 1 (IDO1) inhibitor for immuno-oncology. ACS Med. Chem. Lett. 8, 486–491 (2017).
Mullard, A. IDO takes a blow. Nat. Rev. Drug Discov. 17, 307 (2018).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Hartmann, C. et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 118, 469–474 (2009).
Kang, M. R. et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int. J. Cancer 125, 353–355 (2009).
Sonoda, Y. et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 100, 1996–1998 (2009).
Medeiros, B. C. et al. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia 31, 272 (2016).
Carbonneau, M. et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat. Commun. 7, 12700 (2016).
Popovici-Muller, J. et al. Discovery of AG-120 (ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med. Chem. Lett. 9, 300–305 (2018).
Quivoron, C. et al. AG-221, an oral, selective, first-in-class, potent IDH2-R140Q mutant inhibitor, induces differentiation in a xenotransplant model. Blood 124, 3735 (2014).
DiNardo, C. D. et al. Mutant IDH (mIDH) inhibitors, ivosidenib or enasidenib, with azacitidine (AZA) in patients with acute myeloid leukemia (AML). J. Clin. Oncol. 36, 7042 (2018).
DiNardo, C. D. et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 378, 2386–2398 (2018).
Agarwal, P. et al. Elucidating immunometabolic targets in glioblastoma. Am. J. Cancer Res. 7, 1990–1995 (2017).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018).
Kohanbash, G. et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J. Clin. Invest. 127, 1425–1437 (2017).
Amankulor, N. M. et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 31, 774–786 (2017).
Cordes, T. et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 (2016).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113 (2018).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Weiss, J. M. et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 128, 3794–3805 (2018).
Miller, W. L., Thomas, R. A., Berne, R. M. & Rubio, R. Adenosine production in the ischemic kidney. Circ. Res. 43, 390–397 (1978).
Vijayan, D., Young, A., Teng, M. W. L. & Smyth, M. J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 17, 709–724 (2017).
Antonioli, L., Blandizzi, C., Pacher, P. & Haskó, G. Immunity, inflammation and cancer: a leading role for adenosine. Nat. Rev. Cancer 13, 842–857 (2013).
Cai, X.-Y. et al. High expression of CD39 in gastric cancer reduces patient outcome following radical resection. Oncol. Lett. 12, 4080–4086 (2016).
Lu, X.-X. et al. Expression and clinical significance of CD73 and hypoxia-inducible factor-1α in gastric carcinoma. World J. Gastroenterol. 19, 1912–1918 (2013).
Turcotte, M. et al. CD73 is associated with poor prognosis in high-grade serous ovarian cancer. Cancer Res. 75, 4494 (2015).
Inoue, Y. et al. Prognostic impact of CD73 and A2A adenosine receptor expression in non-small-cell lung cancer. Oncotarget 8, 8738–8751 (2017).
Maj, T. et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 18, 1332–1341 (2017).
Sun, X. et al. CD39/ENTPD1 expression by CD4+Foxp3+ regulatory T cells promotes hepatic metastatic tumor growth in mice. Gastroenterology 139, 1030–1040 (2010).
Zanin, R. F. et al. Differential macrophage activation alters the expression profile of NTPDase and Ecto-5′nucleotidase. PLOS ONE 7, e31205 (2012).
Csóka, B. et al. Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 26, 376–386 (2012).
Huang, S., Apasov, S., Koshiba, M. & Sitkovsky, M. Role of A2a extracellular adenosine receptor-mediated signaling in adenosine-mediated inhibition of T cell activation and expansion. Blood 90, 1600–1610 (1997).
Allard, B., Pommey, S., Smyth, M. J. & Stagg, J. Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin. Cancer Res. 19, 5626–5635 (2013).
Ohta, A. et al. The development and immunosuppressive functions of CD4+ CD25+ FoxP3+ regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Frontiers Immunol. 3, 190 (2012).
Li, L. et al. Dendritic cells tolerized with adenosine A2AR agonist attenuate acute kidney injury. J. Clin. Invest. 122, 3931–3942 (2012).
Sorrentino, C., Miele, L., Porta, A., Pinto, A. & Morello, S. Myeloid-derived suppressor cells contribute to A2B adenosine receptor-induced VEGF production and angiogenesis in a mouse melanoma model. Oncotarget 6, 27478–27489 (2015).
Jackson, S. W. et al. Disordered purinergic signaling inhibits pathological angiogenesis in cd39/Entpd1-null mice. Am. J. Pathol. 171, 1395–1404 (2007).
Merighi, S. et al. Adenosine receptors as mediators of both cell proliferation and cell death of cultured human melanoma cells. J. Invest. Dermatol. 119, 923–933 (2002).
Zhi, X. et al. RNAi-mediated CD73 suppression induces apoptosis and cell-cycle arrest in human breast cancer cells. Cancer Sci. 101, 2561–2569 (2010).
Zhou, P. et al. Overexpression of Ecto-5′-nucleotidase (CD73) promotes T-47D human breast cancer cells invasion and adhesion to extracellular matrix. Cancer Biol. Ther. 6, 426–431 (2007).
Wang, L. et al. Ecto-5′-nucleotidase promotes invasion, migration and adhesion of human breast cancer cells. J. Cancer Res. Clin. Oncol. 134, 365–372 (2008).
Iannone, R., Miele, L., Maiolino, P., Pinto, A. & Morello, S. Adenosine limits the therapeutic effectiveness of anti-CTLA4 mAb in a mouse melanoma model. Am. J. Cancer Res. 4, 172–181 (2014).
Beavis, P. A. et al. Adenosine receptor 2A blockade increases the efficacy of anti–PD-1 through enhanced antitumor T cell responses. Cancer Immunol. Res. 3, 506 (2015).
Mittal, D. et al. Antimetastatic effects of blocking PD-1 and the adenosine A2A receptor. Cancer Res. 74, 3652 (2014).
Corvus Pharmaceuticals. Corvus Pharmaceuticals announces interim results from ongoing phase 1/1b study demonstrating safety and clinical activity of lead checkpoint inhibitor CPI-444 in patients with advanced cancers. GlobeNewsWire https://globenewswire.com/news-release/2017/04/04/954192/0/en/Corvus-Pharmaceuticals-Announces-Interim-Results-from-Ongoing-Phase-1-1b-Study-Demonstrating-Safety-and-Clinical-Activity-of-Lead-Checkpoint-Inhibitor-CPI-444-in-Patients-with-Adva.html (2017).
Martínez-Colón, G. J. & Moore, B. B. Prostaglandin E2 as a regulator of immunity to pathogens. Pharmacol. Ther. 185, 135–146 (2018).
Peebles, R. S. Jr Prostaglandins in asthma and allergic diseases. Pharmacol. Ther. 193, 1–19 (2018).
Furuyashiki, T. & Narumiya, S. Stress responses: the contribution of prostaglandin E(2) and its receptors. Nat. Rev. Endocrinol. 7, 163–175 (2011).
Zelenay, S. et al. Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270 (2015).
Sharma, S. et al. Tumor cyclooxygenase-2/prostaglandin E2-dependent promotion of FOXP3 expression and CD4+ CD25+ T regulatory cell activities in lung cancer. Cancer Res. 65, 5211–5220 (2005).
Baratelli, F. et al. Prostaglandin E2 induces FOXP3 gene expression and T regulatory cell function in human CD4+ T cells. J. Immunol. 175, 1483–1490 (2005).
Mahic, M., Yaqub, S., Johansson, C. C., Taskén, K. & Aandahl, E. M. FOXP3+CD4+CD25+ adaptive regulatory T cells express cyclooxygenase-2 and suppress effector T cells by a prostaglandin E2-dependent mechanism. J. Immunol. 177, 246–254 (2006).
Snijdewint, F. G., Kalinski, P., Wierenga, E. A., Bos, J. D. & Kapsenberg, M. L. Prostaglandin E2 differentially modulates cytokine secretion profiles of human T helper lymphocytes. J. Immunol. 150, 5321–5329 (1993).
Demeure, C. E., Yang, L. P., Desjardins, C., Raynauld, P. & Delespesse, G. Prostaglandin E2 primes naive T cells for the production of anti-inflammatory cytokines. Eur. J. Immunol. 27, 3526–3531 (1997).
Larsson, K. et al. COX/mPGES-1/PGE2 pathway depicts an inflammatory-dependent high-risk neuroblastoma subset. Proc. Natl Acad. Sci. USA 112, 8070–8075 (2015).
Obermajer, N. et al. PGE(2)-driven induction and maintenance of cancer-associated myeloid-derived suppressor cells. Immunol. Invest. 41, 635–657 (2012).
Böttcher, J. P. et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037 (2018).
Chen, J. H. et al. Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nat. Med. 21, 327–334 (2015).
Chia, W. K., Ali, R. & Toh, H. C. Aspirin as adjuvant therapy for colorectal cancer — reinterpreting paradigms. Nat. Rev. Clin. Oncol. 9, 561–570 (2012).
Drew, D. A., Cao, Y. & Chan, A. T. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat. Rev. Cancer 16, 173–186 (2016).
Li, Y. et al. Hydrogel dual delivered celecoxib and anti-PD-1 synergistically improve antitumor immunity. OncoImmunology 5, e1074374 (2016).
Chell, S. D. et al. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res. 66, 3106–3113 (2006).
Buchanan, F. G. et al. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc. Natl Acad. Sci. USA 103, 1492–1497 (2006).
Xu, S. et al. An EP4 antagonist ONO-AE3-208 suppresses cell invasion, migration, and metastasis of prostate cancer. Cell Biochem. Biophys. 70, 521–527 (2014).
Kashiwagi, E. et al. Prostaglandin receptors induce urothelial tumourigenesis as well as bladder cancer progression and cisplatin resistance presumably via modulating PTEN expression. Br. J. Cancer 118, 213–223 (2018).
Majumder, M., Xin, X., Liu, L., Girish, G. V. & Lala, P. K. Prostaglandin E2 receptor EP4 as the common target on cancer cells and macrophages to abolish angiogenesis, lymphangiogenesis, metastasis, and stem-like cell functions. Cancer Sci. 105, 1142–1151 (2014).
Majumder, M. et al. COX-2 induces breast cancer stem cells via EP4/PI3K/AKT/NOTCH/WNT axis. Stem Cells 34, 2290–2305 (2016).
O’Callaghan, G. & Houston, A. Prostaglandin E2 and the EP receptors in malignancy: possible therapeutic targets? Br. J. Pharmacol. 172, 5239–5250 (2015).
Markovic, T., Jakopin, Ž., Dolenc, M. S. & Mlinaric-Rašcan, I. Structural features of subtype-selective EP receptor modulators. Drug Discov. Today 22, 57–71 (2017).
Currie, E., Schulze, A., Zechner, R., Walther, T. C. & Farese, R. V. Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 (2013).
Bochet, L. et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 73, 5657–5668 (2013).
Zhang, Y. et al. Stromal progenitor cells from endogenous adipose tissue contribute to pericytes and adipocytes that populate the tumor microenvironment. Cancer Res. 72, 5198–5208 (2012).
Herber, D. L. et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 16, 880–886 (2010).
Cubillos-Ruiz, J. R. et al. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell 161, 1527–1538 (2015).
Al-Khami, A. A. et al. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology 6, e1344804 (2017).
Niu, Z. et al. Caspase-1 cleaves PPARγ for potentiating the pro-tumor action of TAMs. Nat. Commun. 8, 766 (2017).
Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004 (2018).
McDonald, G. et al. Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J. Clin. Invest. 124, 712–724 (2014).
Bettencourt, I. A. & Powell, J. D. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J. Immunol. 198, 999 (2017).
Zech, T. et al. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 28, 466 (2009).
Owen, D. M. et al. High plasma membrane lipid order imaged at the immunological synapse periphery in live T cells. Mol. Membr. Biol. 27, 178–189 (2010).
Yang, W. et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651 (2016).
Zhang, Y. et al. Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 32, 377–391 (2017).
Chowdhury, P. S., Chamoto, K., Kumar, A. & Honjo, T. PPAR-induced fatty acid oxidation in T cells increases the number of tumor-reactive CD8+ T cells and facilitates anti-PD-1 therapy. Cancer Immunol. Res. 6, 1375–1387 (2018).
Haghikia, A. et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 (2015).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 8, 80 (2014).
York, A. G. et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163, 1716–1729 (2015).
Sag, D., Cekic, C., Wu, R., Linden, J. & Hedrick, C. C. The cholesterol transporter ABCG1 links cholesterol homeostasis and tumour immunity. Nat. Commun. 6, 6354 (2015).
Kannan, Y. et al. TPL-2 regulates macrophage lipid metabolism and M2 differentiation to control TH2-mediated immunopathology. PLOS Pathog. 12, e1005783 (2016).
Huang, S. C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Netea, M. G. et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Mourits, V. P., Wijkmans, J. C., Joosten, L. A. & Netea, M. G. Trained immunity as a novel therapeutic strategy. Curr. Opin. Pharmacol. 41, 52–58 (2018).
Buffen, K. et al. Autophagy controls BCG-induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLOS Pathog. 10, e1004485 (2014).
Cheng, S. C. et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Arts, R. J. et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016).
Freemerman, A. J. et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 (2014).
Patsoukis, N. et al. PD-1 alters T cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Lim, S. et al. Immunoregulatory protein B7-H3 reprograms glucose metabolism in cancer cells by ROS-mediated stabilization of HIF1α. Cancer Res. 76, 2231–2242 (2016).
Parry, R. V. et al. CTLA-4 and PD-1 receptors inhibit T cell activation by distinct mechanisms. Mol. Cell. Biol. 25, 9543–9553 (2005).
Ferris, R. L., Lu, B. & Kane, L. P. Too much of a good thing? Tim-3 and TCR signaling in T cell exhaustion. J. Immunol. 193, 1525–1530 (2014).
Lee, J. et al. Phosphotyrosine-dependent coupling of Tim-3 to T cell receptor signaling pathways. Mol. Cell. Biol. 31, 3963–3974 (2011).
Previte, D. M. et al. Lymphocyte activation gene-3 regulates mitochondrial biogenesis and metabolism of naive CD4+ T cells. J. Immunol. 198 (Suppl), 150.1 (2017).
Jacobs, S. R. et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Klein, G. R. I. et al. Mitochondrial priming by CD28. Cell 171, 385–397 (2017).
Choi, B. K. et al. 4-1BB signaling activates glucose and fatty acid metabolism to enhance CD8+ T cell proliferation. Cell. Mol. Immunol. 14, 748–757 (2017).
Tsurutani, N. et al. Costimulation endows immunotherapeutic CD8 T cells with IL-36 responsiveness during aerobic glycolysis. J. Immunol. 196, 124–134 (2016).
Sabharwal, S. S. et al. GITR agonism enhances cellular metabolism to support CD8+ T cell proliferation and effector cytokine production in a mouse tumor model. Cancer Immunol. Res. 6, 1199–1211 (2018).
Zeng, H. et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45, 540–554 (2016).
Gigoux, M. et al. Inducible costimulator facilitates T-dependent B cell activation by augmenting IL-4 translation. Mol. Immunol. 59, 46–54 (2014).
Menk, A. V. et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 22, 1509–1521 (2018).
Desdin-Mico, G., Soto-Heredero, G. & Mittelbrunn, M. Mitochondrial activity in T cells. Mitochondrion 41, 51–57 (2018).
Sena, L. A. et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013).
Gubser, P. M. et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14, 1064–1072 (2013).
Bantug, G. R. et al. Mitochondria-endoplasmic reticulum contact sites function as immunometabolic hubs that orchestrate the rapid recall response of memory CD8+ T cells. Immunity 48, 542–555 (2018).
Menk, A. V. et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J. Exp. Med. 215, 1091–1100 (2018).
Siska, P. J. et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, 93411 (2017).
Xu, X. et al. Autophagy is essential for effector CD8+ T cell survival and memory formation. Nat. Immunol. 15, 1152–1161 (2014).
Pearce, E. L. et al. Enhancing CD8 T cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Murera, D. et al. CD4 T cell autophagy is integral to memory maintenance. Sci. Rep. 8, 5951 (2018).
Green, D. R., Galluzzi, L. & Kroemer, G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112 (2011).
O’Sullivan, T. E., Johnson, L. R., Kang, H. H. & Sun, J. C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43, 331–342 (2015).
Hinrichs, C. S. & Rosenberg, S. A. Exploiting the curative potential of adoptive T cell therapy for cancer. Immunol. Rev. 257, 56–71 (2014).
Klebanoff, C. A. et al. Determinants of successful CD8+ T cell adoptive immunotherapy for large established tumors in mice. Clin. Cancer Res. 17, 5343–5352 (2011).
Klebanoff, C. A. et al. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc. Natl Acad. Sci. USA 101, 1969–1974 (2004).
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Araki, K. et al. mTOR regulates memory CD8 T cell differentiation. Nature 460, 108–112 (2009).
Crompton, J. G. et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. 75, 296–305 (2015).
Zhang, L. et al. Mammalian target of rapamycin complex 2 controls CD8 T cell memory differentiation in a foxo1-dependent manner. Cell Rep. 14, 1206–1217 (2016).
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016).
Klebanoff, C. A. et al. Inhibition of AKT signaling uncouples T cell differentiation from expansion for receptor-engineered adoptive immunotherapy. JCI Insight 2, 95103 (2017).
Zheng, W. et al. PI3K orchestration of the in vivo persistence of chimeric antigen receptor-modified T cells. Leukemia 32, 1157–1167 (2018).
Frauwirth, K. A. et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Kawalekar, O. U. et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 712 (2016).
de Lima Thomaz, L. et al. The impact of metabolic reprogramming on dendritic cell function. Int. Immunopharmacol. 63, 84–93 (2018).
Acknowledgements
The work of P.R. is supported in part by the Swiss National Science Foundation (CRSII3_160708 and 31003A_156469 grants) and a research grant from Roche Pharma Research and Early Development (pRED). The work of S.C.-C.H. is supported by a Case Comprehensive Cancer Center ASC Pilot Award (IRG-91-022-19). S.-M.F. acknowledges research funding from the European Research Council (ERC) (ERC Consolidator Grant agreement number 771486 — MetaRegulation), the Research Foundation — Flanders (FWO; Odysseus Group II, Research Grants and Research Projects) and KU Leuven (Methuselah Co-Funding). The work of P.-C.H. is supported in part by the Swiss National Science Foundation (31003A_163204 and 31003A_182470 grants), the Melanoma Research Alliance, the Cancer Research Institute (CLIP award), Roche pRED and the Swiss Cancer League (grant KFS-3949-08-2016).
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to researching the data, discussions of content and writing of the manuscript and reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.R. is a member of the scientific advisory board of Immatics and NexImmune and has received speaker honoraria from AstraZeneca, Bristol-Myers Squibb and Roche and research funding from Roche in the form of a Pharma Research and Early Development (pRED) grant. S.-M.F. has received funding from Bayer and Merck. P.-C.H. has received research funding from Idorsia, Novartis and Roche (pRED grant) and speaker honoraria from Chugai and Pfizer and is a member of the scientific advisory board of Elixiron Immunotherapeutics. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, X., Wenes, M., Romero, P. et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol 16, 425–441 (2019). https://doi.org/10.1038/s41571-019-0203-7
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-019-0203-7
This article is cited by
-
Modulating ferroptosis sensitivity: environmental and cellular targets within the tumor microenvironment
Journal of Experimental & Clinical Cancer Research (2024)
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Metabolic reprogramming in the tumor microenvironment of liver cancer
Journal of Hematology & Oncology (2024)
-
Inhibition of autophagy-related protein 7 enhances anti-tumor immune response and improves efficacy of immune checkpoint blockade in microsatellite instability colorectal cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Comprehensive analysis of m6A modification in immune infiltration, metabolism and drug resistance in hepatocellular carcinoma
Cancer Cell International (2024)