Abstract
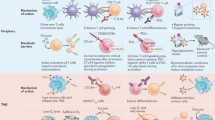
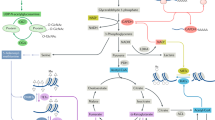
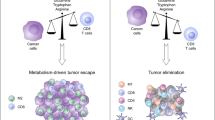
Through the successes of checkpoint blockade and adoptive cellular therapy, immunotherapy has become an established treatment modality for cancer. Cellular metabolism has emerged as a critical determinant of the viability and function of both cancer cells and immune cells. In order to sustain prodigious anabolic needs, tumours employ a specialized metabolism that differs from untransformed somatic cells. This metabolism leads to a tumour microenvironment that is commonly acidic, hypoxic and/or depleted of critical nutrients required by immune cells. In this context, tumour metabolism itself is a checkpoint that can limit immune-mediated tumour destruction. Because our understanding of immune cell metabolism and cancer metabolism has grown significantly in the past decade, we are on the cusp of being able to unravel the interaction of cancer cell metabolism and immune metabolism in therapeutically meaningful ways. Although there are metabolic processes that are seemingly fundamental to both cancer and responding immune cells, metabolic heterogeneity and plasticity may serve to distinguish the two. As such, understanding the differential metabolic requirements of the diverse cells that comprise an immune response to cancer offers an opportunity to selectively regulate immune cell function. Such a nuanced evaluation of cancer and immune metabolism can uncover metabolic vulnerabilities and therapeutic windows upon which to intervene for enhanced immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fox, C. J., Hammerman, P. S. & Thompson, C. B. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5, 844–852 (2005). This paper reviews the critical determinants of metabolic reprogramming that occur during T cell activation, including the roles of co-stimulatory signalling and growth factors, to meet increased bioenergetic demands required for pathogen response.
Andrejeva, G. & Rathmell, J. C. Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab. 26, 49–70 (2017).
Bauer, D. E. et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 18, 1303–1305 (2004).
Kim, J. & DeBerardinis, R. J. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 30, 434–446 (2019).
Weinberg, F. et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA 107, 8788–8793 (2010).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870.e5 (2019).
Chen, P. H. et al. Metabolic diversity in human non-small cell lung cancer cells. Mol. Cell 76, 838–851.e5 (2019).
Cascone, T. et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987.e4 (2018). This study identifies elevated tumour glycolysis as a determinant of immune resistance in melanoma in an adoptive cell therapy model.
Renner, K. et al. Restricting glycolysis preserves T cell effector functions and augments checkpoint therapy. Cell Rep. 29, 135–150.e9 (2019).
Kleffel, S. et al. Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 162, 1242–1256 (2015).
Nunes-Xavier, C. E. et al. Decreased expression of B7-H3 reduces the glycolytic capacity and sensitizes breast cancer cells to AKT/mTOR inhibitors. Oncotarget 7, 6891–6901 (2016).
Lim, S. et al. Immunoregulatory protein B7-H3 reprograms glucose metabolism in cancer cells by ROS-mediated stabilization of HIF1α. Cancer Res. 76, 2231 (2016).
Johnston, R. J. et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature 574, 565–570 (2019).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015). This study demonstrates that glucose consumption by tumours can restrict the glycolytic capacity and IFNγ production of T cells, and that this nutrient competition can be attenuated through checkpoint blockade with antibodies against PD1/PDL1 and CTLA4.
Sharma, N. S. et al. Targeting tumor-intrinsic hexosamine biosynthesis sensitizes pancreatic cancer to anti-PD1 therapy. J. Clin. Investigation 130, 451–465 (2020).
Leone, R. D. et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 (2019). This study demonstrates, using pharmacologic glutamine blockade, the potential of leveraging the ability of CD8+ T cells to use alternative metabolic pathways, including acetate metabolism and glucose anaplerosis, to enhance the antitumour response.
Lukey, M. J., Katt, W. P. & Cerione, R. A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 22, 796–804 (2017).
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). This study demonstrates the role of glucose deprivation within the TME as an novel checkpoint for T cell tumouricidal effector functions. Additionally, the study shows a critical role for the glycolysis metabolite, phosphoenolpyruvate, in sustaining Ca2+-NFAT signalling in activated antitumour T cells.
Fridman, W. H., Zitvogel, L., Sautes-Fridman, C. & Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 (2017).
Tsou, P., Katayama, H., Ostrin, E. J. & Hanash, S. M. The emerging role of B cells in tumor immunity. Cancer Res. 76, 5597–5601 (2016).
Roberts, E. W. et al. Critical role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016).
Jansen, C. S. et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019).
Pardoll, D. Cancer and the immune system: basic concepts and targets for intervention. Semin. Oncol. 42, 523–538 (2015).
Miller, J. F. & Sadelain, M. The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer Cell 27, 439–449 (2015).
Becht, E., Giraldo, N. A., Dieu-Nosjean, M. C., Sautes-Fridman, C. & Fridman, W. H. Cancer immune contexture and immunotherapy. Curr. Opin. Immunol. 39, 7–13 (2016).
Patel, C. H., Leone, R. D., Horton, M. R. & Powell, J. D. Targeting metabolism to regulate immune responses in autoimmunity and cancer. Nat. Rev. Drug Discov. 18, 669–688 (2019).
Menk, A. V. et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 22, 1509–1521 (2018).
Frauwirth, K. A. et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002). This study shows that the upregulated glycolytic rate in activated T cells is dependent on CD28 co-stimulation acting through PI3K–AKT signalling.
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). This study demonstrates that acute metabolic reprogramming in activated T cells, including the upregulation of gylycolytic, pentose phosphate and glutaminolysis pathways, is dependent on the activity of the MYC transcription factor.
Gatza, E. et al. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Sci. Transl Med. 3, 67ra68 (2011).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). This study shows that IFNγ translation in activated T cells is dependent on the upregulation of aerobic glycolysis. The study reports a novel mechanism wherein GAPDH, in the absence of glycolysis-driven NAD+, blocks IFNγ translation through a moonlighting role by binding to the 3′ untranslated region of IFNγ mRNA.
Sena, L. A. et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013).
Pollizzi, K. N. & Powell, J. D. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 14, 435–446 (2014).
Finlay, D. K. et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453 (2012).
Osthus, R. C. et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 275, 21797–21800 (2000).
Patra, K. C. & Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 (2014).
Kidani, Y. et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 14, 489–499 (2013).
Gorrini, C., Harris, I. S. & Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947 (2013).
Hosios, A. M. & Vander Heiden, M. G. The redox requirements of proliferating mammalian cells. J. Biol. Chem. 293, 7490–7498 (2018).
Przybytkowski, E. & Averill-Bates, D. A. Correlation between glutathione and stimulation of the pentose phosphate cyclein situin Chinese hamster ovary cells exposed to hydrogen peroxide. Arch. Biochem. Biophysics 325, 91–98 (1996).
Sena, L. A. & Chandel, N. S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 (2012).
Mak, T. W. et al. Glutathione primes T cell metabolism for inflammation. Immunity 46, 675–689 (2017).
Swamy, M. et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712–720 (2016). This study identifies the regulation of protein O-GlcNAcylation through glucose and glutamine metabolism as a key controller of T cell clonal expansion.
Ma, E. H. et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 25, 345–357 (2017). This study identifies extracellular serine as a key immunometabolite that is required for optimal T cell expansion, in glucose-replete conditions.
Martínez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Blagih, J. et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015).
Cham, C. M. & Gajewski, T. F. Glucose availability regulates IFN-γ production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 174, 4670–4677 (2005).
Cham, C. M., Driessens, G., O’Keefe, J. P. & Gajewski, T. F. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 38, 2438–2450 (2008).
Delgoffe, G. M. et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 (2009).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. investigation 123, 4479–4488 (2013). This study shows that inhibiting glycolysis with 2-DG during in vitro activation enhances the CD8+ T cell memory phenotype and leads to improved antitumour activity after adoptive transfer.
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
Shi, L. Z. et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Dang, E. V. et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011).
Zhao, E. et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 17, 95–103 (2016).
van Bruggen, J. A. C. et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood 134, 44–58 (2019).
Siska, P. J. et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, e93411 (2017).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016).
Nii, T. et al. Molecular events involved in up-regulating human Na+-independent neutral amino acid transporter LAT1 during T-cell activation. Biochem. J. 358, 693–704 (2001).
Carr, E. L. et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 (2010).
Nakaya, M. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 (2014).
Ren, W. et al. Amino-acid transporters in T-cell activation and differentiation. Cell Death Dis. 8, e2655 (2017).
Sinclair, L. V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Geiger, R. et al. l-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e813 (2016). This study shows that elevating l-arginine levels during T cell activation promotes long-lived central memory-like cells with enhanced antitumour activity in a mouse model.
Srivastava, M. K., Sinha, P., Clements, V. K., Rodriguez, P. & Ostrand-Rosenberg, S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 70, 68–77 (2010).
Munn, D. H. et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281, 1191–1193 (1998).
Munn, D. H. et al. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 189, 1363–1372 (1999). This paper uncovers the ability of macrophages to induce cell cycle arrest in Teff cells through the IDO-dependent catabolism of tryptophan.
Munn, D. H. et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642 (2005).
Liu, M. et al. Targeting the IDO1 pathway in cancer: from bench to bedside. J. Hematol. Oncol. 11, 100–100 (2018).
Liu, H. et al. Increased expression of IDO associates with poor postoperative clinical outcome of patients with gastric adenocarcinoma. Sci. Rep. 6, 21319 (2016).
Li, R. et al. IDO inhibits T-cell function through suppressing Vav1 expression and activation. Cancer Biol. Ther. 8, 1402–1408 (2009).
Godin-Ethier, J., Hanafi, L. A., Piccirillo, C. A. & Lapointe, R. Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin. Cancer Res. 17, 6985–6991 (2011).
Cluntun, A. A., Lukey, M. J., Cerione, R. A. & Locasale, J. W. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 3, 169–180 (2017).
Cheng, T. et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl Acad. Sci. USA 108, 8674–8679 (2011).
Nabe, S. et al. Reinforce the antitumor activity of CD8+ T cells via glutamine restriction. Cancer Sci. 109, 3737–3750 (2018).
Reid, M. A., Dai, Z. & Locasale, J. W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19, 1298–1306 (2017).
Yang, W. et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016). This work demonstrates the potential to enhance CD8+ T cell effector function, proliferation and antitumour activity through pharmacologic and genetic blockade of the cholesterol-esterifying enzyme ACAT1, which allows for increased cholesterol content in plasma membranes, facilitating T cell receptor clustering.
Ma, X. et al. Cholesterol induces CD8+ T cell exhaustion in the tumor microenvironment. Cell Metab. 30, 143–156.e5 (2019).
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
van der Windt, G. J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl Acad. Sci. USA 110, 14336–14341 (2013).
Pollizzi, K. N. et al. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Investigation 125, 2090–2108 (2015).
Pollizzi, K. N. et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8+ T cell differentiation. Nat. Immunol. 17, 704–711 (2016).
Raud, B. et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 28, 504–515.e7 (2018).
Pan, Y. et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Chisolm, D. A. et al. CCCTC-binding factor translates interleukin 2- and α-ketoglutarate-sensitive metabolic changes in T cells into context-dependent gene programs. Immunity 47, 251–267.e7 (2017).
Schug, Z. T., Vande Voorde, J. & Gottlieb, E. The metabolic fate of acetate in cancer. Nat. Rev. Cancer 16, 708–717 (2016).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074.e5 (2019).
Balmer, M. L. et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016).
Petrova, V., Annicchiarico-Petruzzelli, M., Melino, G. & Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 7, 10 (2018).
McKeown, S. R. Defining normoxia, physoxia and hypoxia in tumours—implications for treatment response. Br. J. Radiol. 87, 20130676 (2014).
Caldwell, C. C. et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J. Immunol. 167, 6140–6149 (2001).
Ohta, A. et al. In vivo T cell activation in lymphoid tissues is inhibited in the oxygen-poor microenvironment. Front. Immunol. 2, 27 (2011).
Phan, A. T. et al. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016).
Wang, L. et al. CD73 has distinct roles in nonhematopoietic and hematopoietic cells to promote tumor growth in mice. J. Clin. Investigation 121, 2371–2382 (2011).
Allard, B., Longhi, M. S., Robson, S. C. & Stagg, J. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol. Rev. 276, 121–144 (2017).
Ferrante, C. J. et al. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor α (IL-4Rα) signaling. Inflammation 36, 921–931 (2013).
Leone, R. D., Lo, Y. C. & Powell, J. D. A2aR antagonists: next generation checkpoint blockade for cancer immunotherapy. Comput. Struct. Biotechnol. J. 13, 265–272 (2015).
Leone, R. D. & Emens, L. A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 6, 57 (2018).
Waickman, A. T. et al. Enhancement of tumor immunotherapy by deletion of the A2A adenosine receptor. Cancer Immunol. Immunother. 61, 917–926 (2012).
Ohta, A. et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl Acad. Sci. 103, 13132–13137 (2006). This study demonstrates that pharmacologic or genetic inhibition of adenosine-A2AR receptor signalling on T cells enhanced T cell-mediated tumour regression and destruction of metastases.
Leone, R. D. et al. Inhibition of the adenosine A2a receptor modulates expression of T cell coinhibitory receptors and improves effector function for enhanced checkpoint blockade and ACT in murine cancer models. Cancer Immunol. Immunother. 67, 1271–1284 (2018).
Hatfield, S. M. et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci. Transl Med. 7, 277ra230 (2015). This report shows the potential of respiratory hyperoxygenation in mice to suppress adenosine-A2AR signalling in antitumour CD8+ T cells and NK cells, while also weakening Treg cell immunosuppression, leading to enhanced tumour regression and survival.
Fischer, K. et al. Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood 109, 3812–3819 (2007). This study demonstrates the suppressive effect of lactic acid on CD8+ Teff cell proliferation, cytokine production and cytotoxicity.
Brand, A. et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 24, 657–671 (2016).
Mendler, A. N. et al. Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int. J. Cancer 131, 633–640 (2012).
Labadie, B. W., Bao, R. & Luke, J. J. Reimagining IDO Pathway inhibition in cancer immunotherapy via downstream focus on the tryptophan–kynurenine–aryl hydrocarbon axis. Clin. Cancer Res. 25, 1462–1471 (2019).
Liu, Y. et al. Tumor-repopulating cells induce PD-1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell 33, 480–494.e7 (2018).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018). This study shows that tumour-derived R-2-HG is taken up by T cells and interferes with NFAT transcriptional activity and polyamine biosynthesis to suppress T cell activity. The antitumour immune response is enhanced by inhibition of enzymatic activity of IDH1.
Zhang, L. et al. d-2-Hydroxyglutarate is an intercellular mediator in IDH-mutant gliomas inhibiting complement and T cells. Clin. Cancer Res. 24, 5381–5391 (2018).
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016). This study shows that necrosis-related potassium release occurs in human and mouse tumours and that elevated extracellular potassium concentration leads to suppression of Akt–mTOR signalling and T cell function in a PP2A-dependent manner.
Vodnala, S. K. et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, eaau0135 (2019).
Keppel, M. P., Saucier, N., Mah, A. Y., Vogel, T. P. & Cooper, M. A. Activation-specific metabolic requirements for NK cell IFN-γ production. J. Immunol. 194, 1954–1962 (2015).
Assmann, N. et al. Srebp-controlled glucose metabolism is essential for NK cell functional responses. Nat. Immunol. 18, 1197–1206 (2017).
Wu, Q. et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 5, 637–645 (2013).
Rossin, D. et al. Increased production of 27-hydroxycholesterol in human colorectal cancer advanced stage: possible contribution to cancer cell survival and infiltration. Free. Radic. Biol. Med. 136, 35–44 (2019).
Li, D., Long, W., Huang, R., Chen, Y. & Xia, M. 27-Hydroxycholesterol inhibits sterol regulatory element-binding protein 1 activation and hepatic lipid accumulation in mice. Obesity 26, 713–722 (2018).
Guo, F. et al. Upregulation of 24(R/S),25-epoxycholesterol and 27-hydroxycholesterol suppresses the proliferation and migration of gastric cancer cells. Biochem. Biophys. Res. Commun. 504, 892–898 (2018).
Sun, J. C., Ma, A. & Lanier, L. L. Cutting edge: IL-15-independent NK cell response to mouse cytomegalovirus infection. J. Immunol. 183, 2911–2914 (2009).
Cong, J. et al. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab. 28, 243–255.e5 (2018).
Oberlies, J. et al. Regulation of NK cell function by human granulocyte arginase. J. Immunol. 182, 5259–5267 (2009).
Lamas, B. et al. Altered functions of natural killer cells in response to l-arginine availability. Cell Immunol. 280, 182–190 (2012).
Sarkar, S. et al. Hypoxia induced impairment of NK cell cytotoxicity against multiple myeloma can be overcome by IL-2 activation of the NK cells. PLoS One 8, e64835 (2013).
Loftus, R. M. et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun. 9, 2341 (2018).
Balsamo, M. et al. Hypoxia downregulates the expression of activating receptors involved in NK-cell-mediated target cell killing without affecting ADCC. Eur. J. Immunol. 43, 2756–2764 (2013).
Husain, Z., Huang, Y., Seth, P. & Sukhatme, V. P. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells. J. Immunol. 191, 1486–1495 (2013).
Harmon, C. et al. Lactate-mediated acidification of tumor microenvironment induces apoptosis of liver-resident NK cells in colorectal liver metastasis. Cancer Immunol. Res. 7, 335–346 (2019).
Young, A. et al. A2AR adenosine signaling suppresses natural killer cell maturation in the tumor microenvironment. Cancer Res. 78, 1003–1016 (2018).
Orecchioni, M., Ghosheh, Y., Pramod, A. B. & Ley, K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS–) vs. alternatively activated macrophages. Front. Immunol. 10, 1084 (2019).
Martinez, F. O. & Gordon, S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 6, 13 (2014).
Aras, S. & Zaidi, M. R. TAMeless traitors: macrophages in cancer progression and metastasis. Br. J. Cancer 117, 1583–1591 (2017).
Freemerman, A. J. et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 (2014).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Haschemi, A. et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826 (2012).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Iles, K. E. & Forman, H. J. Macrophage signaling and respiratory burst. Immunol. Res. 26, 95–105 (2002).
Viola, A., Munari, F., Sanchez-Rodriguez, R., Scolaro, T. & Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019).
Zajac, E. et al. Angiogenic capacity of M1- and M2-polarized macrophages is determined by the levels of TIMP-1 complexed with their secreted proMMP-9. Blood 122, 4054–4067 (2013).
Stuehr, D. J. & Nathan, C. F. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 169, 1543–1555 (1989).
El-Gayar, S., Thuring-Nahler, H., Pfeilschifter, J., Rollinghoff, M. & Bogdan, C. Translational control of inducible nitric oxide synthase by IL-13 and arginine availability in inflammatory macrophages. J. Immunol. 171, 4561–4568 (2003).
Moon, J. S. et al. mTORC1-induced HK1-dependent glycolysis regulates NLRP3 inflammasome activation. Cell Rep. 12, 102–115 (2015).
Jantsch, J. et al. Hypoxia and hypoxia-inducible factor-1α modulate lipopolysaccharide-induced dendritic cell activation and function. J. Immunol. 180, 4697–4705 (2008).
Krawczyk, C. M. et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 (2010).
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785 (2007).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Gerriets, V. A. et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Investigation 125, 194–207 (2015).
Pacella, I. et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc. Natl Acad. Sci. USA 115, E6546–e6555 (2018).
Cluxton, D., Petrasca, A., Moran, B. & Fletcher, J. M. Differential regulation of human Treg and TH17 cells by fatty acid synthesis and glycolysis. Front. Immunol. 10, 115 (2019).
Sun, I. H. et al. mTOR complex 1 signaling regulates the generation and function of central and effector Foxp3+ regulatory T cells. J. Immunol. 201, 481–492 (2018).
Angelin, A. et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e87 (2017). This study shows that the Treg cell-defining transcription factor, FOXP3, suppresses MYC activity and glycolysis, while increasing OXPHOS, allowing Treg cells to resist lactate-mediated suppression in low-glucose environments such as the TME.
Araujo, L., Khim, P., Mkhikian, H., Mortales, C. L. & Demetriou, M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. eLife 6, e21330 (2017).
Klysz, D. et al. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal. 8, ra97 (2015).
Facciabene, A. et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature 475, 226–230 (2011).
Clambey, E. T. et al. Hypoxia-inducible factor-1α-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl Acad. Sci. USA 109, E2784–E2793 (2012).
Westendorf, A. M. et al. Hypoxia enhances immunosuppression by inhibiting CD4+ effector T cell function and promoting Treg activity. Cell Physiol. Biochem. 41, 1271–1284 (2017).
Ben-Shoshan, J., Maysel-Auslender, S., Mor, A., Keren, G. & George, J. Hypoxia controls CD4+CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1α. Eur. J. Immunol. 38, 2412–2418 (2008).
Miska, J. et al. HIF-1α is a metabolic switch between glycolytic-driven migration and oxidative phosphorylation-driven immunosuppression of Tregs in glioblastoma. Cell Rep. 27, 226–237.e4 (2019).
Ohta, A. & Sitkovsky, M. Extracellular adenosine-mediated modulation of regulatory T cells. Front. Immunol. 5, 304 (2014).
Ohta, A. et al. The development and immunosuppressive functions of CD4+CD25+FoxP3+ regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front. Immunol. 3, 190 (2012).
Lee, J. H., Elly, C., Park, Y. & Liu, Y. C. E3 ubiquitin ligase VHL regulates hypoxia-inducible factor-1α to maintain regulatory T cell stability and suppressive capacity. Immunity 42, 1062–1074 (2015).
Fallarino, F. et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor ζ-chain and induce a regulatory phenotype in naive T cells. J. Immunol. 176, 6752–6761 (2006).
Chen, W., Liang, X., Peterson, A. J., Munn, D. H. & Blazar, B. R. The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J. Immunol. 181, 5396–5404 (2008).
Mezrich, J. D. et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190 (2010).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Divakaruni, A. S. et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 28, 490–503.e7 (2018).
Shearer, J. D., Richards, J. R., Mills, C. D. & Caldwell, M. D. Differential regulation of macrophage arginine metabolism: a proposed role in wound healing. Am. J. Physiol. 272, E181–E190 (1997).
Ye, C. et al. Targeting ornithine decarboxylase by α-difluoromethylornithine inhibits tumor growth by impairing myeloid-derived suppressor cells. J. Immunol. 196, 915 (2016).
Mills, C. D., Shearer, J., Evans, R. & Caldwell, M. D. Macrophage arginine metabolism and the inhibition or stimulation of cancer. J. Immunol. 149, 2709–2714 (1992).
Hayes, C. S. et al. Polyamine-blocking therapy reverses immunosuppression in the tumor microenvironment. Cancer Immunol. Res. 2, 274–285 (2014).
Hossain, F. et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol. Res. 3, 1236–1247 (2015).
Jian, S. L. et al. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death Dis. 8, e2779 (2017).
Casazza, A. et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 24, 695–709 (2013).
Murdoch, C. & Lewis, C. E. Macrophage migration and gene expression in response to tumor hypoxia. Int. J. Cancer 117, 701–708 (2005).
Henze, A. T. & Mazzone, M. The impact of hypoxia on tumor-associated macrophages. J. Clin. Investigation 126, 3672–3679 (2016).
Tripathi, C. et al. Macrophages are recruited to hypoxic tumor areas and acquire a pro-angiogenic M2-polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget 5, 5350–5368 (2014).
Csóka, B. et al. Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 26, 376–386 (2012).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). This study demonstrates that lactic acid from tumour cells induces an M2-like phenotype in macrophages with upregulation of vascular endothelial growth factor and ARG1.
Li, W. et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 28, 87–103.e6 (2018).
Corzo, C. A. et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 207, 2439–2453 (2010).
Crompton, J. G. et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. 75, 296–305 (2015). This work shows that harvested tumour infiltrating lymphocytes expanded in the presence of an inhibitor of AKT show enhanced transcriptional, metabolic and functional properties of Tmem cells, imbuing these T cells with enhanced in vivo persistence and augmented antitumor activity in mouse models.
He, W. et al. CD155T/TIGIT signaling regulates CD8+ T-cell metabolism and promotes tumor progression in human gastric cancer. Cancer Res. 77, 6375–6388 (2017).
Sabharwal, S. S. et al. GITR agonism enhances cellular metabolism to support CD8+ T-cell proliferation and effector cytokine production in a mouse tumor model. Cancer Immunol. Res. 6, 1199 (2018).
Parry, R. V. et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell Biol. 25, 9543–9553 (2005).
Schaer, D. A. et al. The folate pathway inhibitor pemetrexed pleiotropically enhances effects of cancer immunotherapy. Clin. Cancer Res. 25, 7175–7188 (2019). This study demonstrates that pemetrexed augments antitumour immunity in combination with anti-PDL1 checkpoint blockade in mouse models, in part by enhancing CD8+ T cell metabolic health through stimulating mitochondrial biogenesis with subsequent increased T cell infiltration and activation.
Sousa, C. M. et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016).
Zhang, W. et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat. Cell Biol. 14, 276–286 (2012).
Ko, Y.-H. et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol. Ther. 12, 1085–1097 (2011).
Zhang, Z. et al. Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat. Med. 24, 617–627 (2018).
Okano, T. et al. 3-Bromopyruvate ameliorate autoimmune arthritis by modulating TH17/Treg cell differentiation and suppressing dendritic cell activation. Sci. Rep. 7, 42412 (2017).
Telang, S. et al. Discovery of a PFKFB3 inhibitor for phase I trial testing that synergizes with the B-Raf inhibitor vemurafenib. Cancer Metab. 2, P14–P14 (2014).
Kornberg, M. D. et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02546440 (2020).
Liberti, M. V. et al. A predictive model for selective targeting of the Warburg effect through GAPDH inhibition with a natural product. Cell Metab. 26, 648–659.e8 (2017).
Dunbar, E. M. et al. Phase 1 trial of dichloroacetate (DCA) in adults with recurrent malignant brain tumors. Invest. N. Drugs 32, 452–464 (2014).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01386632 (2020).
Yeung, C. et al. Targeting glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma. Cancer Res. 79, 5060–5073 (2019).
Holubarsch, ChristianJ. F. et al. A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (Etomoxir for the Recovery of Glucose Oxidation) study. Clin. Sci. 113, 205–212 (2007).
Senanayake, E. L. et al. Multicentre double-blind randomized controlled trial of perhexiline as a metabolic modulator to augment myocardial protection in patients with left ventricular hypertrophy undergoing cardiac surgery. Eur. J. Cardiothorac. Surg. 48, 354–362 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00602199 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00061646 (2020).
Alkhouri, N., Lawitz, E., Noureddin, M., DeFronzo, R. & Shulman, G. I. GS-0976 (Firsocostat): an investigational liver-directed acetyl-CoA carboxylase (ACC) inhibitor for the treatment of non-alcoholic steatohepatitis (NASH). Expert. Opin. Investig. Drugs 29, 135–141 (2020).
Meuwese, M. C. et al. ACAT inhibition and progression of carotid atherosclerosis in patients with familial hypercholesterolemia: the CAPTIVATE randomized trial. Jama 301, 1131–1139 (2009).
Scharping, N. E., Menk, A. V., Whetstone, R. D., Zeng, X. & Delgoffe, G. M. Efficacy of PD-1 blockade Is potentiated by metformin-induced reduction of tumor hypoxia. Cancer immunology Res. 5, 9–16 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03048500 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03800602 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03994744 (2020).
Xiang, Y. et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Investigation 125, 2293–2306 (2015).
Gross, M. I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Therapeutics 13, 890–901 (2014).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04265534 (2020).
Lee, C. F. et al. Preventing allograft rejection by targeting immune metabolism. Cell Rep. 13, 760–770 (2015).
Adjei, A. A. Pemetrexed (ALIMTA), A. Novel multitargeted antineoplastic agent. Clin. Cancer Res. 10, 4276s (2004).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03793179 (2020).
Chabner, B. A., Myers, C. E., Coleman, C. N. & Johns, D. G. The clinical pharmacology of antineoplastic agents (first of two parts). N. Engl. J. Med. 292, 1107–1113 (1975).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00989352 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03643276 (2020).
Longley, D. B., Harkin, D. P. & Johnston, P. G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 (2003).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02997228 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03777813 (2020).
Suwannaroj, S., Lagoo, A., Keisler, D. & McMurray, R. W. Antioxidants suppress mortality in the female NZB × NZW F1 mouse model of systemic lupus erythematosus (SLE). Lupus 10, 258–265 (2001).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00003346 (2020).
Yang, Z. et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci. Transl Med. 8, 331ra338 (2016).
Mele, L. et al. A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 9, 572 (2018).
Ricciardiello, F. et al. Inhibition of the hexosamine biosynthetic pathway by targeting PGM3 causes breast cancer growth arrest and apoptosis. Cell Death Dis. 9, 377 (2018).
Steggerda, S. M. et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J. Immunother. Cancer 5, 101 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03361228 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02903914 (2020).
Przystal, J. M. et al. Efficacy of arginine depletion by ADI-PEG20 in an intracranial model of GBM. Cell Death Dis. 9, 1192 (2018).
Prendergast, G. C., Malachowski, W. P., DuHadaway, J. B. & Muller, A. J. Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 77, 6795–6811 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03493945 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04231864 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04049669 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02073123 (2020).
Megias-Vericat, J. E., Ballesta-Lopez, O., Barragan, E. & Montesinos, P. IDH1-mutated relapsed or refractory AML: current challenges and future prospects. Blood Lymphat. Cancer 9, 19–32 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03684811 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02719574 (2020).
Li, X. et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 16, 425–441 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03742102 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04262388 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02754141 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02403193 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03337698 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03381274 (2020).
Vander Heiden, M. G. & DeBerardinis, R. J. Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017).
Saeidi, A. et al. T-cell exhaustion in chronic infections: reversing the state of exhaustion and reinvigorating optimal protective immune responses. Front. Immunol. 9, 2569 (2018).
Bengsch, B. et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 (2016). This paper reports the suppression of glycolysis, mitochondrial dysfunction and mitochondrial biogenesis in CD8+ T cells through PD1 signalling leading to an exhausted T cell phenotype.
Teijeira, A. et al. Mitochondrial morphological and functional reprogramming following CD137 (4-1BB) costimulation. Cancer Immunol. Res. 6, 798 (2018).
Menk, A. V. et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J. Exp. Med. 215, 1091 (2018).
Zhao, Z. et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell 28, 415–428 (2015).
Milone, M. C. et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 17, 1453–1464 (2009).
Imai, C. et al. Chimeric receptors with 4-1BB signaling capacity provoke potent cytotoxicity against acute lymphoblastic leukemia. Leukemia 18, 676–684 (2004).
Finney, H. M., Akbar, A. N. & Lawson, A. D. Activation of resting human primary T cells with chimeric receptors: costimulation from CD28, inducible costimulator, CD134, and CD137 in series with signals from the TCRζ chain. J. Immunol. 172, 104–113 (2004).
Davila, M. L. et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl Med. 6, 224ra225 (2014).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
J.D.P. is a scientific founder, a paid consultant and has equity in Dracen Pharmaceuticals. Technology arising in part from the studies described herein was patented by Johns Hopkins University and subsequently licensed to Dracen Pharmaceuticals (JHU083 is currently labelled as DRP-083). R.D.L. and J.D.P. are inventors for pending patent application no. PCT/US16/44829 submitted by Johns Hopkins University that covers the use of glutamine analogues, such as JHU083 (DRP-083), for cancer immunotherapy. J.D.P has been a paid consultant for Corvus Pharmaceuticals and has equity in the company.
Additional information
Peer review information
Nature Reviews Cancer thanks N. Chandel, J. Fan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Oxidative phosphorylation
-
(OXPHOS). A highly efficient form of cellular respiration synthesizing ATP from the phosphorylation of ADP using electrochemical potential energy generated by the transfer of electrons from NADH or FADH2 to oxygen through a series of mitochondrial electron carriers.
- Immune checkpoint pathways
-
Pathways mediated by cell surface proteins on immune cells, such as PD1 or CTLA4, that serve to suppress the immune response, which can be activated by ligands within the tumour microenvironment or draining lymph nodes.
- Chimeric antigen receptor T cells
-
(CAR T cells). T cells harvested from a patient’s blood and genetically modified to express a special receptor that can recognize and respond to specific, predefined molecular targets on tumour cells.
- Hexosamine biosynthesis pathway
-
(HBP). A branch of glycolysis that generates building blocks used for glycosylation of proteins and lipids.
- Pentose phosphate pathway
-
(PPP). A metabolic branch of glycolysis generating NADPH, used for fatty acid synthesis and redox homeostasis, and 5-carbon sugars used in nucleotide synthesis.
- Nuclear factor of activated T cells
-
(NFAT). A calcium-dependent transcription factor activated in response to T cell receptor stimulation. Cooperation with the AP-1 transcription factor results in a productive immune response and transcription of pro-inflammatory cytokines, such as IL-2 and interferon-γ.
- Cataplerosis
-
The loss of metabolic intermediates in a metabolic pathway (particularly the tricarboxylic acid cycle) owing to consumption or degradation.
- Anaplerosis
-
The process of replenishing intermediates of the tricarboxylic acid cycle to support biosynthesis.
- De novo lipid synthesis
-
The cellular biosynthesis of fatty acids, triglycerides, cholesterol and other lipids from carbohydrates or other non-lipid precursors.
- 2-Oxoglutarate-dependent dioxygenases
-
(2OGDD). A family of enzymes that catalyse the hydroxylation of macromolecules, often as a prerequisite to demethylation, reliant on α-ketoglutarate, Fe2+, ascorbate and oxygen as cofactors.
- Major histocompatibility complex
-
(MHC). MHC class I (MHC-I) is expressed on all nucleated cells, a molecular complex presenting intracellular peptide epitopes for CD8+ T cell receptor recognition. Also expressed on antigen-presenting cells, allowing initial antigen-specific activation of cytotoxic CD8+ T cells. MHC-II is highly expressed on antigen-presenting cells for presenting antigenic epitopes for CD4+ T cell receptor recognition and activation.
- Antigen cross-presentation
-
The ability of antigen-presenting cells to process extracellular antigens and present them to CD8+ T cells through major histocompatibility complex class 1 presentation.
Rights and permissions
About this article
Cite this article
Leone, R.D., Powell, J.D. Metabolism of immune cells in cancer. Nat Rev Cancer 20, 516–531 (2020). https://doi.org/10.1038/s41568-020-0273-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-020-0273-y
This article is cited by
-
Metabolic reprogramming in the tumor microenvironment of liver cancer
Journal of Hematology & Oncology (2024)
-
Metabolic regulation of tumor-associated macrophage heterogeneity: insights into the tumor microenvironment and immunotherapeutic opportunities
Biomarker Research (2024)
-
Targeting the macrophage immunocheckpoint: a novel insight into solid tumor immunotherapy
Cell Communication and Signaling (2024)
-
A glutamine tug-of-war between cancer and immune cells: recent advances in unraveling the ongoing battle
Journal of Experimental & Clinical Cancer Research (2024)
-
Fatty acid metabolism of immune cells: a new target of tumour immunotherapy
Cell Death Discovery (2024)