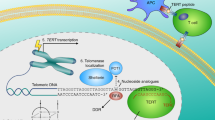
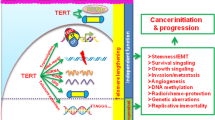
Abstract
Cancer cells establish replicative immortality by activating a telomere-maintenance mechanism (TMM), be it telomerase or the alternative lengthening of telomeres (ALT) pathway. Targeting telomere maintenance represents an intriguing opportunity to treat the vast majority of all cancer types. Whilst telomerase inhibitors have historically been heralded as promising anticancer agents, the reality has been more challenging, and there are currently no therapeutic options for cancer types that use ALT despite their aggressive nature and poor prognosis. In this Review, we discuss the mechanistic differences between telomere maintenance by telomerase and ALT, the current methods used to detect each mechanism, the utility of these tests for clinical diagnosis, and recent developments in the therapeutic strategies being employed to target both telomerase and ALT. We present notable developments in repurposing established therapeutic agents and new avenues that are emerging to target cancer types according to which TMM they employ. These opportunities extend beyond inhibition of telomere maintenance, by finding and exploiting inherent weaknesses in the telomeres themselves to trigger rapid cellular effects that lead to cell death.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). This paper identifies the key hallmarks of cancer, one of which is infinite replicative potential that can be enabled by activation of a TMM.
Guterres, A. N. & Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 39, 5811–5824 (2020).
Rousseau, P. & Autexier, C. Telomere biology: rationale for diagnostics and therapeutics in cancer. RNA Biol. 12, 1078–1082 (2015).
Meyne, J., Ratliff, R. L. & Moyzis, R. K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl Acad. Sci. USA 86, 7049–7053 (1989).
Harley, C. B., Futcher, A. B. & Greider, C. W. Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990).
Griffith, J. D. et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999).
Larrivee, M., LeBel, C. & Wellinger, R. J. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18, 1391–1396 (2004).
Makarov, V. L., Hirose, Y. & Langmore, J. P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666 (1997).
Wright, W. E., Tesmer, V. M., Huffman, K. E., Levene, S. D. & Shay, J. W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11, 2801–2809 (1997).
Erdel, F. et al. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 18, 41–53 (2017).
Chong, L. et al. A human telomeric protein. Science 270, 1663–1667 (1995).
de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005).
Van Ly, D. et al. Telomere loop dynamics in chromosome end protection. Mol. Cell 71, 510–525.e6 (2018).
Tomaska, L., Nosek, J., Kar, A., Willcox, S. & Griffith, J. D. A new view of the T-loop junction: implications for self-primed telomere extension, expansion of disease-related nucleotide repeat blocks, and telomere evolution. Front. Genet. https://doi.org/10.3389/fgene.2019.00792 (2019).
Ghanim, G. E. et al. Structure of human telomerase holoenzyme with bound telomeric DNA. Nature 593, 449–453 (2021). This paper shows sub-4 Å resolution of the human telomerase holoenzyme bound to telomeric DNA.
Zhang, Q., Kim, N.-K. & Feigon, J. Architecture of human telomerase RNA. Proc. Natl Acad. Sci. USA 108, 20325–20332 (2011).
Zhang, J.-M. & Zou, L. Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci. 10, 30 (2020).
Sobinoff, A. P. & Pickett, H. A. Mechanisms that drive telomere maintenance and recombination in human cancers. Curr. Opin. Genet. Dev. 60, 25–30 (2020). This paper provides an in-depth review of the current understanding of the ALT mechanism.
Jafri, M. A., Ansari, S. A., Alqahtani, M. H. & Shay, J. W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 8, 69 (2016).
Vulliamy, T. et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA 105, 8073–8078 (2008).
Pogacic, V., Dragon, F. & Filipowicz, W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell Biol. 20, 9028–9040 (2000).
Cerone, M. A., Ward, R. J., Londono-Vallejo, J. A. & Autexier, C. Telomerase RNA mutated in autosomal dyskeratosis congenita reconstitutes a weakly active telomerase enzyme defective in telomere elongation. Cell Cycle 4, 585–589 (2005).
Zhong, F. L. et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012).
Nandakumar, J. et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012).
Chen, L. Y., Redon, S. & Lingner, J. The human CST complex is a terminator of telomerase activity. Nature 488, 540–544 (2012).
Latrick, C. M. & Cech, T. R. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29, 924–933 (2010).
Kelleher, C., Kurth, I. & Lingner, J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell Biol. 25, 808–818 (2005).
Smogorzewska, A. & de Lange, T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 (2004).
Ye, J. Z. & de Lange, T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 36, 618–623 (2004).
Lee, S. S., Bohrson, C., Pike, A. M., Wheelan, S. J. & Greider, C. W. ATM kinase is required for telomere elongation in mouse and human cells. Cell Rep. 13, 1623–1632 (2015).
Leteurtre, F., Li, X., Gluckman, E. & Carosella, E. D. Telomerase activity during the cell cycle and in gamma-irradiated hematopoietic cells. Leukemia 11, 1681–1689 (1997).
Schmidt, J. C., Zaug, A. J. & Cech, T. R. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.e9 (2016).
Sarek, G., Vannier, J. B., Panier, S., Petrini, J. H. J. & Boulton, S. J. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell 57, 622–635 (2015).
Zhao, Y. et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475 (2009).
Barthel, F. P. et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 49, 349–357 (2017).
Hiyama, E. & Hiyama, K. Telomere and telomerase in stem cells. Br. J. Cancer 96, 1020–1024 (2007).
Blasco, M. A., Funk, W., Villeponteau, B. & Greider, C. W. Functional characterisation and developmental regulation of mouse telomerase RNA. Science 269, 1267–1270 (1995).
Zhang, F., Cheng, D., Wang, S. & Zhu, J. Human specific regulation of the telomerase reverse transcriptase gene. Genes 7, 30 (2016).
Sieverling, L. et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 11, 733 (2020).
de Nonneville, A. & Reddel, R. R. Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX. Nat. Commun. 12, 1552 (2021). This paper provides a comprehensive analysis of the prevalence of ATRX and/or DAXX mutations across cancer types.
Wang, S., Hu, C. & Zhu, J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol. Biol. Cell 18, 669–677 (2007).
Wang, S. & Zhu, J. The hTERT gene is embedded in a nuclease-resistant chromatin domain. J. Biol. Chem. 279, 55401–55410 (2004).
Stern, J. L., Theodorescu, D., Vogelstein, B., Papadopoulos, N. & Cech, T. R. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 29, 2219–2224 (2015).
Episkopou, H. et al. Alternative lengthening of telomeres is characterised by reduced compaction of telomeric chromatin. Nucleic Acids Res. 42, 4391–4405 (2014).
Napier, C. E. et al. ATRX represses alternative lengthening of telomeres. Oncotarget 6, 16543–16558 (2015).
Clynes, D. et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538 (2015).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localisation at specific genomic regions. Cell 140, 678–691 (2010).
Lewis, P. W., Elsaesser, S. J., Noh, K. M., Stadler, S. C. & Allis, C. D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010).
Li, F. et al. ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalisation. EMBO J. 38, e96659 (2019).
Lee, M. et al. Telomere sequence content can be used to determine ALT activity in tumours. Nucleic Acids Res. 46, 4903–4918 (2018). This study provides the first demonstration that telomere variant repeat content can be used to accurately identify ALT-positive tumours from whole-genome sequencing data.
Gauchier, M. et al. SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 5, eaav3673 (2019).
Conomos, D., Reddel, R. R. & Pickett, H. A. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 21, 760–770 (2014). This paper demonstrates that NuRD–ZNF827 is recruited exclusively to ALT-positive telomeres, where it functions to remodel telomeric chromatin, creating a recombination-permissive environment that enables ALT activity.
Gaillard, H., García-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Sfeir, A. et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103 (2009).
Vannier, J.-B., Pavicic-Kaltenbrunner, V., Petalcorin, M. I. R., Ding, H. & Boulton, S. J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149, 795–806 (2012).
Cesare, A. J. et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 16, 1244–1251 (2009).
Déjardin, J. & Kingston, R. E. Purification of proteins associated with specific genomic loci. Cell 136, 175–186 (2009).
Garcia-Exposito, L. et al. Proteomic profiling reveals a specific role for translesion DNA polymerase η in the alternative lengthening of telomeres. Cell Rep. 17, 1858–1871 (2016).
Conomos, D. et al. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 199, 893–906 (2012). This study shows that telomere variant repeats become interspersed throughout ALT-positive telomeres, thereby disrupting shelterin binding.
Cox, K. E., Maréchal, A. & Flynn, R. L. SMARCAL1 resolves replication stress at ALT telomeres. Cell Rep. 14, 1032–1040 (2016).
Lu, R. et al. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 10, 2252 (2019).
Root, H. et al. FANCD2 limits BLM-dependent telomere instability in the alternative lengthening of telomeres pathway. Hum. Mol. Genet. 25, 3255–3268 (2016).
Silva, B. et al. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 10, 2253 (2019).
Pan, X., Ahmed, N., Kong, J. & Zhang, D. Breaking the end: target the replication stress response at the ALT telomeres for cancer therapy. Mol. Cell. Oncol. 4, e1360978 (2017).
Pan, X. et al. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 9, 19110 (2019). Along with Lu et al.61 and Silva et al.63, this study demonstrates the role of the FANCM–BTR complex in alleviating replication stress at ALT-positive telomeres and provides the first evidence for the FANCM–BTR interaction as a novel drug target.
Pan, X. et al. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl Acad. Sci. USA 114, E5940–E5949 (2017).
Betous, R. et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26, 151–162 (2012).
Poole, L. A. et al. SMARCAL1 maintains telomere integrity during DNA replication. Proc. Natl Acad. Sci. USA 112, 14864–14869 (2015).
Dilley, R. L. et al. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58 (2016). This paper provides a definition of the mechanism of break-induced telomere synthesis at ALT-positive telomeres, and its reliance on the unique RFC–PCNA–Polδ replisome.
Nimonkar, A. V., Özsoy, A. Z., Genschel, J., Modrich, P. & Kowalczykowski, S. C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl Acad. Sci. USA 105, 16906–16911 (2008).
Nimonkar, A. V. et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 (2011).
Zhang, J.-M., Yadav, T., Ouyang, J., Lan, L. & Zou, L. Alternative lengthening of telomeres through two distinct break-induced replication pathways. Cell Rep. 26, 955–968.e3 (2019).
Min, J., Wright, W. E. & Shay, J. W. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell. Biol. 37, e00226-17 (2017).
Min, J., Wright, W. E. & Shay, J. W. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33, 814–827 (2019).
Verma, P. et al. RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev. 33, 221–235 (2019).
Sobinoff, A. P. & Pickett, H. A. Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet. 33, 921–932 (2017).
Costantino, L. et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 (2014).
Sobinoff, A. P. et al. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 36, 2907–2919 (2017). This study demonstrates that BLM and SLX4 have opposing functions at ALT-positive telomeres, with BLM promoting telomere extension and SLX4 counteracting telomere extension by inducing telomere exchange events.
Wu, L. & Hickson, I. D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003). This paper demonstrates that BLM works together with TOP3A to promote the dissolution of Holliday junctions.
Svendsen, J. M. et al. Mammalian BTBD12/SLX4 assembles a holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 (2009).
Müller, S., Matunis, M. J. & Dejean, A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17, 61–70 (1998).
Chung, I., Leonhardt, H. & Rippe, K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 124, 3603–3618 (2011).
Henson, J. D. et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 (2009).
Zhang, J. M., Genois, M. M., Ouyang, J., Lan, L. & Zou, L. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol. Cell 81, 1027–1042.e4 (2021).
Lauer, N. K. et al. Absence of telomerase activity in malignant bone tumors and soft-tissue sarcomas. Sarcoma 6, 43–46 (2002).
Odago, F. O. & Gerson, S. L. Telomerase inhibition and telomere erosion: a two-pronged strategy in cancer therapy. Trends Pharmacol. Sci. 24, 328–331 (2003).
Mo, Y. Q. et al. Simultaneous targeting of telomeres and telomerase as a cancer therapeutic approach. Cancer Res. 63, 579–585 (2003).
Lee, H. W. et al. Essential role of mouse telomerase in highly proliferative organs. Nature 392, 569–574 (1998).
Flores, I., Cayuela, M. L. & Blasco, M. A. Effects of telomerase and telomere length on epidermal stem cell behavior. Science 309, 1253–1256 (2005).
Hernandez-Sanchez, W. et al. A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity. PLoS Biol. 17, e3000204 (2019).
Gryaznov, S. et al. Telomerase inhibitors- oligonucleotide phosphoramidates as potential therapeutic agents. Nucleosides Nucleotides Nucleic Acids 20, 401–410 (2001).
Chen, Z., Monia, B. P. & Corey, D. R. Telomerase inhibition, telomere shortening, and decreased cell proliferation by cell permeable 2′-O-methoxyethyl oligonucleotides. J. Med. Chem. 45, 5423–5425 (2002).
Herbert, B. S., Pongracz, K., Shay, J. W. & Gryaznov, S. M. Oligonucleotide N3′–>P5′ phosphoramidates as efficient telomerase inhibitors. Oncogene 21, 638–642 (2002).
Pitts, A. E. & Corey, D. R. Inhibition of human telomerase by 2′-O-methyl-RNA. Proc. Natl Acad. Sci. USA 95, 11549–11554 (1998).
Schrank, Z. et al. Oligonucleotides targeting telomeres and telomerase in cancer. Molecules https://doi.org/10.3390/molecules23092267 (2018).
Eckburg, A., Dein, J., Berei, J., Schrank, Z. & Puri, N. Oligonucleotides and microRNAs targeting telomerase subunits in cancer therapy. Cancers https://doi.org/10.3390/cancers12092337 (2020).
Ferrandon, S. et al. Telomerase inhibition improves tumor response to radiotherapy in a murine orthotopic model of human glioblastoma. Mol. Cancer 14, 134 (2015).
Marian, C. O. et al. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin. Cancer Res. 16, 154–163 (2010).
Frink, R. E. et al. Telomerase inhibitor imetelstat has preclinical activity across the spectrum of non-small cell lung cancer oncogenotypes in a telomere length dependent manner. Oncotarget 7, 31639–31651 (2016).
Salloum, R. et al. A molecular biology and phase II study of imetelstat (GRN163L) in children with recurrent or refractory central nervous system malignancies: a pediatric brain tumor consortium study. J. Neurooncol. 129, 443–451 (2016).
Barwe, S. P., Huang, F., Kolb, E. A. & Gopalakrishnapillai, A. Imetelstat significantly reduces leukemia stem cells in patient-derived xenograft models of pediatric AML. Blood 138, 3352 (2021).
Steensma, D. P. et al. Imetelstat achieves meaningful and durable transfusion independence in high transfusion-burden patients with lower-risk myelodysplastic syndromes in a phase II study. J. Clin. Oncol. 39, 48–56 (2021).
Tefferi, A. et al. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N. Engl. J. Med. 373, 908–919 (2015).
Baerlocher, G. M. et al. Telomerase inhibitor imetelstat in patients with essential thrombocythemia. N. Engl. J. Med. 373, 920–928 (2015). This study demonstrates the success of imetelstat in treating patients with thrombocythaemia.
Dikmen, Z. G. et al. In vivo inhibition of lung cancer by GRN163L: a novel human telomerase inhibitor. Cancer Res. 65, 7866–7873 (2005).
Saygin, C. & Carraway, H. E. Current and emerging strategies for management of myelodysplastic syndromes. Blood Rev. 48, 100791 (2021).
Kuykendall, A. T. et al. Favorable overall survival with imetelstat in relapsed/refractory myelofibrosis patients compared with real-world data. Ann. Hematol. 101, 139–146 (2022).
Wang, X. et al. Imetelstat, a telomerase inhibitor, is capable of depleting myelofibrosis stem and progenitor cells. Blood Adv. 2, 2378–2388 (2018).
Thompson, P. A. et al. A phase I trial of imetelstat in children with refractory or recurrent solid tumors: a Children’s Oncology Group Phase I Consortium Study (ADVL1112). Clin. Cancer Res. 19, 6578–6584 (2013).
Chiappori, A. A. et al. A randomised phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 26, 354–362 (2015).
Gomez, D. E., Armando, R. G. & Alonso, D. F. AZT as a telomerase inhibitor. Front. Oncol. 2, 113 (2012).
Leão, R. et al. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J. Biomed. Sci. 25, 22 (2018).
Peng, Y., Mian, I. S. & Lue, N. F. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7, 1201–1211 (2001).
Sanford, S. L., Welfer, G. A., Freudenthal, B. D. & Opresko, P. L. Mechanisms of telomerase inhibition by oxidised and therapeutic dNTPs. Nat. Commun. 11, 5288 (2020).
Mitsuya, H. et al. 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc. Natl Acad. Sci. USA 82, 7096–7100 (1985).
Strahl, C. & Blackburn, E. H. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalised human cell lines. Mol. Cell Biol. 16, 53–65 (1996).
Strahl, C. & Blackburn, E. H. The effects of nucleoside analogs on telomerase and telomeres in Tetrahymena. Nucleic Acids Res. 22, 893–900 (1994).
Wang, Y., Gallagher-Jones, M., Sušac, L., Song, H. & Feigon, J. A structurally conserved human and Tetrahymena telomerase catalytic core. Proc. Natl Acad. Sci. USA 117, 31078–31087 (2020).
Datta, A. et al. Persistent inhibition of telomerase reprograms adult T-cell leukemia to p53-dependent senescence. Blood 108, 1021–1029 (2006).
Langford, A., Ruf, B., Kunze, R., Pohle, H. D. & Reichart, P. Regression of oral Kaposi’s sarcoma in a case of AIDS on Zidovudine (AZT). Br. J. Dermatol. 120, 709–713 (1989).
Lee, R. K. et al. Azidothymidine and interferon-α induce apoptosis in herpesvirus-associated lymphomas1. Cancer Res. 59, 5514–5520 (1999).
Wang, H., Zhou, J., He, Q., Dong, Y. & Liu, Y. Azidothymidine inhibits cell growth and telomerase activity and induces DNA damage in human esophageal cancer. Mol. Med. Rep. 15, 4055–4060 (2017).
Rha, S. Y. et al. Effect of telomere and telomerase interactive agents on human tumor and normal cell lines. Clin. Cancer Res. 6, 987–993 (2000).
Sengupta, S. et al. Induced telomere damage to treat telomerase expressing therapy-resistant pediatric brain tumors. Mol. Cancer Ther. 17, 1504–1514 (2018).
Mender, I., Gryaznov, S., Dikmen, Z. G., Wright, W. E. & Shay, J. W. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2′-deoxyguanosine. Cancer Discov. 5, 82–95 (2015).
Damm, K. et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 20, 6958–6968 (2001).
Rao, Y. K., Kao, T. Y., Wu, M. F., Ko, J. L. & Tzeng, Y. M. Identification of small molecule inhibitors of telomerase activity through transcriptional regulation of hTERT and calcium induction pathway in human lung adenocarcinoma A549 cells. Bioorg. Med. Chem. 18, 6987–6994 (2010).
Yang, Y. L. et al. Histone deacetylase inhibitor AR42 regulates telomerase activity in human glioma cells via an Akt-dependent mechanism. Biochem. Biophys. Res. Commun. 435, 107–112 (2013).
Li, Y. et al. A small molecule compound IX inhibits telomere and attenuates oncogenesis of drug-resistant leukemia cells. FASEB J. 34, 8843–8857 (2020).
Bryan, C. et al. Structural basis of telomerase inhibition by the highly specific BIBR1532. Structure 23, 1934–1942 (2015).
Pascolo, E. et al. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 277, 15566–15572 (2002).
El-Daly, H. et al. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood 105, 1742–1749 (2005).
Giunco, S. et al. Anti-proliferative and pro-apoptotic effects of short-term inhibition of telomerase in vivo and in human malignant B cells xenografted in zebrafish. Cancers 12, 2052 (2020).
Pourbagheri-Sigaroodi, A. et al. Contributory role of microRNAs in anti-cancer effects of small molecule inhibitor of telomerase (BIBR1532) on acute promyelocytic leukemia cell line. Eur. J. Pharmacol. 846, 49–62 (2019).
Kong, W. et al. Knockdown of hTERT and treatment with BIBR1532 inhibit cell proliferation and invasion in endometrial cancer cells. J. Cancer 6, 1337–1345 (2015).
Kusoglu, A., Goker Bagca, B., Ozates Ay, N. P., Gunduz, C. & Biray Avci, C. Telomerase inhibition regulates EMT mechanism in breast cancer stem cells. Gene 759, 145001 (2020).
Biray Avci, C. et al. Effects of telomerase inhibitor on epigenetic chromatin modification enzymes in malignancies. J. Cell. Biochem. 119, 9817–9824 (2018).
Doğan, F. et al. Investigation of the effect of telomerase inhibitor BIBR1532 on breast cancer and breast cancer stem cells. J. Cell. Biochem. https://doi.org/10.1002/jcb.27089 (2018).
Ward, R. J. & Autexier, C. Pharmacological telomerase inhibition can sensitise drug-resistant and drug-sensitive cells to chemotherapeutic treatment. Mol. Pharmacol. 68, 779–786 (2005).
Seimiya, H. et al. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol. Cancer Ther. 1, 657–665 (2002).
Fang, M. Z. et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 63, 7563–7570 (2003).
Berletch, J. B. et al. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J. Cell Biochem. 103, 509–519 (2008).
Naasani, I., Seimiya, H. & Tsuruo, T. Telomerase inhibition, telomere shortening, and senescence of cancer cells by tea catechins. Biochem. Biophys. Res. Commun. 249, 391–396 (1998).
Gurung, R. L., Lim, S. N., Low, G. K. & Hande, M. P. MST-312 Alters telomere dynamics, gene expression profiles and growth in human breast cancer cells. J. Nutrigenet. Nutrigenomics 7, 283–298 (2014).
Fujiwara, C. et al. Cell-based chemical fingerprinting identifies telomeres and lamin A as modifiers of DNA damage response in cancer cells. Sci. Rep. 8, 14827 (2018).
Andrade da Mota, T. H. et al. Effects of in vitro short- and long-term treatment with telomerase inhibitor in U-251 glioma cells. Tumour Biol. 43, 327–340 (2021).
Ghasemimehr, N., Farsinejad, A., Mirzaee Khalilabadi, R., Yazdani, Z. & Fatemi, A. The telomerase inhibitor MST-312 synergistically enhances the apoptotic effect of doxorubicin in pre-B acute lymphoblastic leukemia cells. Biomed. Pharmacother. 106, 1742–1750 (2018).
Zhou, C., Gehrig, P. A., Whang, Y. E. & Boggess, J. F. Rapamycin inhibits telomerase activity by decreasing the hTERT mRNA level in endometrial cancer cells. Mol. Cancer Ther. 2, 789–795 (2003).
Betori, R. C. et al. Targeted covalent inhibition of telomerase. ACS Chem. Biol. 15, 706–717 (2020).
Zhang, N. et al. Bufalin inhibits hTERT expression and colorectal cancer cell growth by targeting CPSF4. Cell Physiol. Biochem. 40, 1559–1569 (2016).
Mizushina, Y., Takeuchi, T., Sugawara, F. & Yoshida, H. Anti-cancer targeting telomerase inhibitors: β-rubromycin and oleic acid. Mini Rev. Med. Chem. 12, 1135–1143 (2012).
Ueno, T. et al. Inhibition of human telomerase by rubromycins: implication of spiroketal system of the compounds as an active moiety. Biochemistry 39, 5995–6002 (2000).
Ellingsen, E. B., Mangsbo, S. M., Hovig, E. & Gaudernack, G. Telomerase as a target for therapeutic cancer vaccines and considerations for optimizing their clinical potential. Front. Immunol. 12, 682492 (2021).
Negrini, S., De Palma, R. & Filaci, G. Anti-cancer immunotherapies targeting telomerase. Cancers 12, 2260 (2020).
Dosset, M., Castro, A., Carter, H. & Zanetti, M. Telomerase and CD4 T cell immunity in cancer. Cancers 12, 1687 (2020).
Bernhardt, S. L. et al. Telomerase peptide vaccination of patients with non-resectable pancreatic cancer: a dose escalating phase I/II study. Br. J. Cancer 95, 1474–1482 (2006).
Schlapbach, C., Yerly, D., Daubner, B., Yawalkar, N. & Hunger, R. E. Telomerase-specific GV1001 peptide vaccination fails to induce objective tumor response in patients with cutaneous T cell lymphoma. J. Dermatol. Sci. 62, 75–83 (2011).
Staff, C., Mozaffari, F., Frödin, J. E., Mellstedt, H. & Liljefors, M. Telomerase (GV1001) vaccination together with gemcitabine in advanced pancreatic cancer patients. Int. J. Oncol. 45, 1293–1303 (2014).
Greten, T. F. et al. A phase II open label trial evaluating safety and efficacy of a telomerase peptide vaccination in patients with advanced hepatocellular carcinoma. BMC Cancer 10, 209 (2010).
Middleton, G. et al. Gemcitabine and capecitabine with or without telomerase peptide vaccine GV1001 in patients with locally advanced or metastatic pancreatic cancer (TeloVac): an open-label, randomised, phase 3 trial. Lancet Oncol. 15, 829–840 (2014).
Kyte, J. A. et al. Telomerase peptide vaccination combined with temozolomide: a clinical trial in stage IV melanoma patients. Clin. Cancer Res. 17, 4568–4580 (2011).
van der Burg, S. H. Correlates of immune and clinical activity of novel cancer vaccines. Semin. Immunol. 39, 119–136 (2018).
Inderberg-Suso, E. M., Trachsel, S., Lislerud, K., Rasmussen, A. M. & Gaudernack, G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. Oncoimmunology 1, 670–686 (2012).
Lilleby, W. et al. Phase I/IIa clinical trial of a novel hTERT peptide vaccine in men with metastatic hormone-naive prostate cancer. Cancer Immunol. Immunother. 66, 891–901 (2017).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT04382664 (2020).
Gridelli, C. et al. Clinical activity of a htert (vx-001) cancer vaccine as post-chemotherapy maintenance immunotherapy in patients with stage IV non-small cell lung cancer: final results of a randomised phase 2 clinical trial. Br. J. Cancer 122, 1461–1466 (2020).
Vetsika, E. K. et al. Immunological responses in cancer patients after vaccination with the therapeutic telomerase-specific vaccine Vx-001. Cancer Immunol. Immunother. 61, 157–168 (2012).
Brunsvig, P. F. et al. Long-term outcomes of a phase I study with UV1, a second generation telomerase based vaccine, in patients with advanced non-small cell lung cancer. Front. Immunol. 11, 572172 (2020).
Thalmensi, J. et al. Anticancer DNA vaccine based on human telomerase reverse transcriptase generates a strong and specific T cell immune response. Oncoimmunology 5, e1083670 (2015).
Teixeira, L. et al. A first-in-human phase I study of INVAC-1, an optimised human telomerase DNA vaccine in patients with advanced solid tumors. Clin. Cancer Res. 26, 588–597 (2020).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT03265717 (2017).
Hwang, H. et al. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure 22, 842–853 (2014).
Zahler, A. M., Williamson, J. R., Cech, T. R. & Prescott, D. M. Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720 (1991).
Zaug, A. J., Podell, E. R. & Cech, T. R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA 102, 10864–10869 (2005).
Wang, H., Nora, G. J., Ghodke, H. & Opresko, P. L. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem. 286, 7479–7489 (2011).
Chaires, J. B. et al. Human POT1 unfolds G-quadruplexes by conformational selection. Nucleic Acids Res. 48, 4976–4991 (2020).
De Cian, A. et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl Acad. Sci. USA 104, 17347–17352 (2007).
Kim, M. Y., Vankayalapati, H., Shin-Ya, K., Wierzba, K. & Hurley, L. H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc. 124, 2098–2099 (2002).
Binz, N., Shalaby, T., Rivera, P., Shin-ya, K. & Grotzer, M. A. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur. J. Cancer 41, 2873–2881 (2005).
Shammas, M. A. et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin. Cancer Res. 10, 770–776 (2004).
Tauchi, T. et al. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene 25, 5719–5725 (2006).
Grand, C. L. et al. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 1, 565–573 (2002).
Mikami-Terao, Y. et al. Antitumor activity of TMPyP4 interacting G-quadruplex in retinoblastoma cell lines. Exp. Eye Res. 89, 200–208 (2009).
Huppert, J. L. & Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 (2005).
Moye, A. L. et al. Telomeric G-quadruplexes are a substrate and site of localisation for human telomerase. Nat. Commun. 6, 7643 (2015).
Oganesian, L., Moon, I. K., Bryan, T. M. & Jarstfer, M. B. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 25, 1148–1159 (2006).
Paudel, B. P. et al. A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. eLife 9, e56428 (2020).
Cimino-Reale, G. et al. miR-380-5p-mediated repression of TEP1 and TSPYL5 interferes with telomerase activity and favours the emergence of an “ALT-like” phenotype in diffuse malignant peritoneal mesothelioma cells. J. Hematol. Oncol. 10, 140 (2017).
Bechter, O. E., Zou, Y., Walker, W., Wright, W. E. & Shay, J. W. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 64, 3444–3451 (2004).
Graham, M. K. et al. Functional loss of ATRX and TERC activates alternative lengthening of telomeres (ALT) in LAPC4 prostate cancer cells. Mol. Cancer Res. 17, 2480–2491 (2019).
Hu, J. et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663 (2012).
Henson, J. D., Neumann, A. A., Yeager, T. R. & Reddel, R. R. Alternative lengthening of telomeres in mammalian cells. Oncogene 21, 598–610 (2002).
Recagni, M., Bidzinska, J., Zaffaroni, N. & Folini, M. The role of alternative lengthening of telomeres mechanism in cancer: translational and therapeutic implications. Cancers https://doi.org/10.3390/cancers12040949 (2020).
Gocha, A. R., Nuovo, G., Iwenofu, O. H. & Groden, J. Human sarcomas are mosaic for telomerase-dependent and telomerase-independent telomere maintenance mechanisms: implications for telomere-based therapies. Am. J. Pathol. 182, 41–48 (2013).
Henson, J. D. & Reddel, R. R. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 584, 3800–3811 (2010).
Matsuo, T. et al. Alternative lengthening of telomeres as a prognostic factor in malignant fibrous histiocytomas of bone. Anticancer Res. 30, 4959–4962 (2010).
Lawlor, R. T. et al. Alternative lengthening of telomeres (ALT) influences survival in soft tissue sarcomas: a systematic review with meta-analysis. BMC Cancer 19, 232 (2019).
Wang, Y., Luo, W. & Wang, Y. PARP-1 and its associated nucleases in DNA damage response. DNA Repair 81, 102651 (2019).
Caron, M. C. et al. Poly(ADP-ribose) polymerase-1 antagonises DNA resection at double-strand breaks. Nat. Commun. 10, 2954 (2019).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Ronson, G. E. et al. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 9, 746 (2018).
Ray Chaudhuri, A. & Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017).
Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012).
Pommier, Y., O’Connor, M. J. & de Bono, J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps317 (2016).
Hopkins, T. A. et al. PARP1 trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol. Cancer Res. 17, 409–419 (2019).
Zimmermann, M. et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018).
Demény, M. A. & Virág, L. The PARP enzyme family and the hallmarks of cancer part 1. Cell intrinsic hallmarks. Cancers https://doi.org/10.3390/cancers13092042 (2021).
Mukherjee, J. et al. A subset of PARP inhibitors induces lethal telomere fusion in ALT-dependent tumor cells. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.abc7211 (2021).
Mukherjee, J. et al. Mutant IDH1 cooperates with ATRX loss to drive the alternative lengthening of telomere phenotype in glioma. Cancer Res. 78, 2966–2977 (2018).
Principe, D. R. Precision medicine for BRCA/PALB2-mutated pancreatic cancer and emerging strategies to improve therapeutic responses to PARP inhibition. Cancers https://doi.org/10.3390/cancers14040897 (2022).
Wu, K. et al. Evaluation of the efficacy of PARP inhibitors in metastatic castration-resistant prostate cancer: a systematic review and meta-analysis. Front. Pharmacol. 12, 777663 (2021).
Cortesi, L., Rugo, H. S. & Jackisch, C. An overview of PARP inhibitors for the treatment of breast cancer. Target. Oncol. 16, 255–282 (2021).
Chiang, Y. C., Lin, P. H. & Cheng, W. F. Homologous recombination deficiency assays in epithelial ovarian cancer: current status and future direction. Front. Oncol. 11, 675972 (2021).
Kamel, D., Gray, C., Walia, J. S. & Kumar, V. PARP inhibitor drugs in the treatment of breast, ovarian, prostate and pancreatic cancers: an update of clinical trials. Curr. Drug Targets 19, 21–37 (2018).
Ma, W., He, H. & Wang, H. Oncolytic herpes simplex virus and immunotherapy. BMC Immunol. 19, 40 (2018).
Andtbacka, R. H. I. et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 33, 2780–2788 (2015).
De Clercq, E. Antiviral drugs in current clinical use. J. Clin. Virol. 30, 115–133 (2004).
Sokolowski, N. A., Rizos, H. & Diefenbach, R. J. Oncolytic virotherapy using herpes simplex virus: how far have we come? Oncolytic Virother. 4, 207–219 (2015).
Liu, T. C. et al. Dominant-negative fibroblast growth factor receptor expression enhances antitumoral potency of oncolytic herpes simplex virus in neural tumors. Clin. Cancer Res. 12, 6791–6799 (2006).
Lukashchuk, V. & Everett, R. D. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84, 4026–4040 (2010).
Everett, R. D., Parada, C., Gripon, P., Sirma, H. & Orr, A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82, 2661–2672 (2008).
Poon, A. P., Liang, Y. & Roizman, B. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 77, 12671–12678 (2003).
Han, M. et al. Synthetic lethality of cytolytic HSV-1 in cancer cells with ATRX and PML deficiency. J. Cell. Sci. https://doi.org/10.1242/jcs.222349 (2019). This paper identifies a mutant version of HSV1 that can be used to selectively induce lysis in ALT-positive cancer cells.
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02588105 (2015).
Koneru, B. et al. ALT neuroblastoma chemoresistance due to telomere dysfunction-induced ATM activation is reversible with ATM inhibitor AZD0156. Sci. Transl. Med. 13, eabd5750 (2021).
Forment, J. V. & O’Connor, M. J. Targeting the replication stress response in cancer. Pharmacol. Ther. 188, 155–167 (2018).
Byun, T. S., Pacek, M., Yee, M. C., Walter, J. C. & Cimprich, K. A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes. Dev. 19, 1040–1052 (2005).
Delacroix, S., Wagner, J. M., Kobayashi, M., Yamamoto, K. & Karnitz, L. M. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21, 1472–1477 (2007).
Gorecki, L., Andrs, M., Rezacova, M. & Korabecny, J. Discovery of ATR kinase inhibitor berzosertib (VX-970, M6620): clinical candidate for cancer therapy. Pharmacol. Ther. 210, 107518 (2020).
Barnieh, F. M., Loadman, P. M. & Falconer, R. A. Progress towards a clinically-successful ATR inhibitor for cancer therapy. Curr. Res. Pharmacol. Drug Discov. 2, 100017 (2021).
Huntoon, C. J. et al. ATR inhibition broadly sensitises ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 73, 3683–3691 (2013).
Kurmasheva, R. T. et al. Initial testing (stage 1) of M6620 (formerly VX-970), a novel ATR inhibitor, alone and combined with cisplatin and melphalan, by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer https://doi.org/10.1002/pbc.26825 (2018).
Flynn, R. L. et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 347, 273–277 (2015). This paper shows the potential efficacy of ATR inhibitors on ALT-associated cells and has led to the progression of these agents into clinical trials for patients with ALT-positive cancers.
Deeg, K. I., Chung, I., Bauer, C. & Rippe, K. Cancer cells with alternative lengthening of telomeres do not display a general hypersensitivity to ATR inhibition. Front. Oncol. 6, 186 (2016).
Laroche-Clary, A. et al. ATR inhibition broadly sensitises soft-tissue sarcoma cells to chemotherapy independent of alternative lengthening telomere (ALT) status. Sci. Rep. 10, 7488 (2020).
Southgate, H. E. D., Chen, L., Tweddle, D. A. & Curtin, N. J. ATR inhibition potentiates PARP inhibitor cytotoxicity in high risk neuroblastoma cell lines by multiple mechanisms. Cancers https://doi.org/10.3390/cancers12051095 (2020).
Yazinski, S. A. et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017).
Kim, H. et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat. Commun. 11, 3726 (2020).
Lloyd, R. L. et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene 39, 4869–4883 (2020).
Yap, T. A. et al. A first-in-human phase I study of ATR inhibitor M1774 in patients with solid tumors. J. Clin. Oncol. 39, TPS3153 (2021).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT04170153 (2019).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT04267939 (2022).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT04497116 (2020).
Schwab, R. A. et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 60, 351–361 (2015).
Huang, J. et al. Remodeling of interstrand crosslink proximal replisomes is dependent on ATR, FANCM, and FANCD2. Cell Rep. 27, 1794–1808.e5 (2019).
Gari, K., Décaillet, C., Stasiak, A. Z., Stasiak, A. & Constantinou, A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29, 141–148 (2008).
Voter, A. F., Manthei, K. A. & Keck, J. L. A high-throughput screening strategy to identify protein-protein interaction inhibitors that block the Fanconi anemia DNA repair pathway. J. Biomol. Screen. 21, 626–633 (2016).
Bryan, T. M. G-Quadruplexes at telomeres: friend or foe? Molecules 25, 3686 (2020).
Tsai, Y. C. et al. A G-quadruplex stabiliser induces M-phase cell cycle arrest. J. Biol. Chem. 284, 22535–22543 (2009).
Amato, R. et al. G-quadruplex stabilisation fuels the ALT pathway in ALT-positive Osteosarcoma cells. Genes 11, 304 (2020).
Pennarun, G. et al. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 24, 2917–2928 (2005).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Woo, S.-R., Corrales, L. & Gajewski, T. F. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 36, 250–256 (2015).
Chen, Y. A. et al. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol. 24, 1124–1131 (2017).
Chen, Q. et al. Apo2L/TRAIL and Bcl-2-related proteins regulate type I interferon-induced apoptosis in multiple myeloma. Blood 98, 2183–2192 (2001).
Chawla-Sarkar, M., Leaman, D. W. & Borden, E. C. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin. Cancer Res. 7, 1821–1831 (2001).
Sanceau, J., Poupon, M. F., Delattre, O., Sastre-Garau, X. & Wietzerbin, J. Strong inhibition of Ewing tumor xenograft growth by combination of human interferon-alpha or interferon-beta with ifosfamide. Oncogene 21, 7700–7709 (2002).
Naka, T. et al. Effects of tumor necrosis factor-related apoptosis-inducing ligand alone and in combination with chemotherapeutic agents on patients’ colon tumors grown in SCID mice. Cancer Res. 62, 5800–5806 (2002).
Thai, L. M. et al. Apo2l/Tumor necrosis factor-related apoptosis-inducing ligand prevents breast cancer-induced bone destruction in a mouse model. Cancer Res. 66, 5363–5370 (2006).
Singh, T. R., Shankar, S., Chen, X., Asim, M. & Srivastava, R. K. Synergistic interactions of chemotherapeutic drugs and tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand on apoptosis and on regression of breast carcinoma in vivo. Cancer Res. 63, 5390–5400 (2003).
Naik, S., Nace, R., Barber, G. N. & Russell, S. J. Potent systemic therapy of multiple myeloma utilizing oncolytic vesicular stomatitis virus coding for interferon-β. Cancer Gene Ther. 19, 443–450 (2012).
Potts, P. R. & Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14, 581–590 (2007).
Kim, Y. S., Keyser, S. G. L. & Schneekloth, J. S. Jr. Synthesis of 2′,3′,4′-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg. Med. Chem. Lett. 24, 1094–1097 (2014).
Zhou, P. et al. 2-D08 as a SUMOylation inhibitor induced ROS accumulation mediates apoptosis of acute myeloid leukemia cells possibly through the deSUMOylation of NOX2. Biochem. Biophys. Res. Commun. 513, 1063–1069 (2019).
Cheng, X. & Kao, H.-Y. Post-translational modifications of PML: consequences and implications. Front. Oncol. https://doi.org/10.3389/fonc.2012.00210 (2013).
Episkopou, H., Diman, A., Claude, E., Viceconte, N. & Decottignies, A. TSPYL5 depletion induces specific death of ALT cells through USP7-dependent proteasomal degradation of POT1. Mol. Cell 75, 469–482.e6 (2019). This study demonstrates that components of APBs protect shelterin complex components from degradation and are rational therapeutic targets.
Diotti, R. & Loayza, D. Shelterin complex and associated factors at human telomeres. Nucleus 2, 119–135 (2011).
Zimmermann, M., Kibe, T., Kabir, S. & de Lange, T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 28, 2477–2491 (2014).
Li, X. et al. Dynamics of TRF1 organizing a single human telomere. Nucleic Acids Res. 49, 760–775 (2020).
Bejarano, L. et al. Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell 32, 590–607.e4 (2017).
Bejarano, L. et al. Multiple cancer pathways regulate telomere protection. EMBO Mol. Med. 11, e10292 (2019).
García-Beccaria, M. et al. Therapeutic inhibition of TRF1 impairs the growth of p53-deficient K-RasG12V-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 7, 930–949 (2015).
Méndez-Pertuz, M. et al. Modulation of telomere protection by the PI3K/AKT pathway. Nat. Commun. 8, 1278 (2017).
Benarroch-Popivker, D. et al. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell 61, 274–286 (2016).
Rai, R., Chen, Y., Lei, M. & Chang, S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun. 7, 10881 (2016).
Ran, X. et al. Design of high-affinity stapled peptides to target the repressor activator protein 1 (RAP1)/telomeric repeat-binding factor 2 (TRF2) protein-protein interaction in the shelterin complex. J. Med. Chem. 59, 328–334 (2016).
Chen, X. et al. Cyclic peptidic mimetics of apollo peptides targeting telomeric repeat binding factor 2 (TRF2) and apollo interaction. ACS Med. Chem. Lett. 9, 507–511 (2018).
Di Maro, S. et al. Shading the TRF2 recruiting function: a new horizon in drug development. J. Am. Chem. Soc. 136, 16708–16711 (2014).
MacKenzie, D. Jr. et al. ALT positivity in human cancers: prevalence and clinical insights. Cancers 13, 2384 (2021).
Hájek, M., Matulová, N., Votruba, I., Holý, A. & Tloust’ová, E. Inhibition of human telomerase by diphosphates of acyclic nucleoside phosphonates. Biochem. Pharmacol. 70, 894–900 (2005).
De Clercq, E. Tanovea® for the treatment of lymphoma in dogs. Biochem. Pharmacol. 154, 265–269 (2018).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02854072 (2016).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT01935154 (2013).
Cummaro, A., Fotticchia, I., Franceschin, M., Giancola, C. & Petraccone, L. Binding properties of human telomeric quadruplex multimers: a new route for drug design. Biochimie 93, 1392–1400 (2011).
Mulholland, K., Siddiquei, F. & Wu, C. Binding modes and pathway of RHPS4 to human telomeric G-quadruplex and duplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Phys. Chem. Chem. Phys. 19, 18685–18694 (2017).
Di Antonio, M., Rodriguez, R. & Balasubramanian, S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods 57, 84–92 (2012).
Nguyen, T. H. D. et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195 (2018).
Dagg, R. A. et al. Extensive proliferation of human cancer cells with ever-shorter telomeres. Cell Rep. 19, 2544–2556 (2017).
Viceconte, N. et al. Highly aggressive metastatic melanoma cells unable to maintain telomere length. Cell Rep. 19, 2529–2543 (2017).
Hartlieb, S. A. et al. Alternative lengthening of telomeres in childhood neuroblastoma from genome to proteome. Nat. Commun. 12, 1269 (2021).
Claude, E. et al. Detection of alternative lengthening of telomeres mechanism on tumor sections. Mol. Biomed. 2, 32 (2021).
Heaphy, C. M. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011).
Lau, L. M. et al. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 41, e34 (2013).
Henson, J. D. et al. The C-circle assay for alternative-lengthening-of-telomeres activity. Methods 114, 74–84 (2017).
Hayward, N. K. et al. Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017).
Lovejoy, C. A. et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 (2012). This study uses fibroblast cell lines with the same genetic background but different TMMs to directly compare the effects of telomerase versus ALT positivity upon cell biology.
Kaul, Z. et al. Functional characterisation of miR-708 microRNA in telomerase positive and negative human cancer cells. Sci. Rep. 11, 17052 (2021).
Wright, W. E., Pereira-Smith, O. M. & Shay, J. W. Reversible cellular senescence: implications for immortalisation of normal human diploid fibroblasts. Mol. Cell. Biol. 9, 3088–3092 (1989).
Tilman, G. et al. Subtelomeric DNA hypomethylation is not required for telomeric sister chromatid exchanges in ALT cells. Oncogene 28, 1682–1693 (2009).
Perrem, K., Colgin, L. M., Neumann, A. A., Yeager, T. R. & Reddel, R. R. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21, 3862–3875 (2001).
Marie-Egyptienne, D. T., Brault, M. E., Zhu, S. & Autexier, C. Telomerase inhibition in a mouse cell line with long telomeres leads to rapid telomerase reactivation. Exp. Cell Res. 314, 668–675 (2008).
Liu, W. et al. Kras mutations increase telomerase activity and targeting telomerase is a promising therapeutic strategy for Kras-mutant NSCLC. Oncotarget 8, 179–190 (2017).
Hu, Y., Bobb, D., Lu, Y., He, J. & Dome, J. S. Effect of telomerase inhibition on preclinical models of malignant rhabdoid tumor. Cancer Genet. 207, 403–411 (2014).
Li, Q. et al. Human telomerase reverse transcriptase as a therapeutic target of dihydroartemisinin for esophageal squamous cancer. Front. Pharmacol. 12, 769787 (2021).
Frank, L. et al. ALT-FISH quantifies alternative lengthening of telomeres activity by imaging of single-stranded repeats. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac113 (2022). Along with Claude et al.292, this study documents a new method to accurately identify ALT-positive cells using native telomere-FISH.
Tejera, A. M., Alonso, D. F., Gomez, D. E. & Olivero, O. A. Chronic in vitro exposure to 3′-azido-2′,3′-dideoxythymidine induces senescence and apoptosis and reduces tumorigenicity of metastatic mouse mammary tumor cells. Breast Cancer Res. Treat. 65, 93–99 (2001).
Vera, E., Bernardes de Jesus, B., Foronda, M., Flores, J. M. & Blasco, M. A. The rate of increase of short telomeres predicts longevity in mammals. Cell Rep. 2, 732–737 (2012).
Lauvrak, S. U. et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer 109, 2228–2236 (2013).
Mohseny, A. B. et al. Functional characterisation of osteosarcoma cell lines provides representative models to study the human disease. Lab. Invest. 91, 1195–1205 (2011).
Acknowledgements
The authors acknowledge the Australian Medical Research Future Fund (2007488) for funding. The authors thank Alexander Sobinoff, Robert Lu and Robyn Yeh for proofreading the manuscript.
Author information
Authors and Affiliations
Contributions
J.G. researched data for the article. J.G. and H.A.P. contributed substantially to discussion of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
H.A.P. is a co-founder and shareholder of Tessellate Bio. J.G. declares no competing interests.
Peer review
Peer review information
Nature Reviews Cancer thanks Lifeng Xu, Vinay Tergaonkar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Homology-directed repair
-
(HDR). A type of double-strand break repair where a homologous section on a sister chromatid is used as a template to guide DNA synthesis and repair. It involves processing of the double-strand break by the MRN complex to create single-stranded overhangs, prior to RAD51-mediated or RAD52-mediated strand invasion of the sister chromatid to enable DNA extension. Intermediates are then resolved to complete the repair.
- Rolling circle amplification reaction
-
An isothermal DNA or RNA amplification reaction where circular oligonucleotides (for example, C-circles) function as a template for the DNA or RNA polymerase.
- 5′ Resection
-
A process where the blunt end of a double-strand break undergoes nucleolytic degradation in the 5′ to 3′ direction to leave a 3′ single-stranded overhang.
- Telomere replication
-
Replication of the telomere repeat tracks.
- C-strand fill-in
-
Telomeres consist of G-strand (5′-TTAGGG-3′) and complementary C-strand (3′-AATCCC-5′) repeats. Telomerase extends the G-strand, resulting in a G-rich single-stranded 3′ overhang. C-strand fill-in is the process by which the complementary C-strand is synthesized by DNA polymerase α-primase 12 to convert the single-stranded DNA of the 3′ overhang into double-stranded DNA.
- G-quadruplexes
-
(G4s). Non-canonical secondary structures formed by guanine (G)-rich DNA sequences.
- Displacement-loops
-
(D-loops). D-loops form when single-stranded DNA invades a section of double-stranded DNA, causing it to separate into a loop structure.
- Strand invasion
-
Single-stranded DNA invades a section of double-stranded DNA with sequence homology.
- Osteoid
-
An unmineralized organic tissue that becomes calcified and contributes to the bone matrix.
- Replisome
-
A protein complex that can exhibit helicase, primase and DNA polymerase activities to replicate DNA of both the leading and lagging strand. During ALT, the replisome consists of proliferating cell nuclear antigen (PCNA), replication factor C (RFC) and DNA polymerase δ (Polδ).
- Branch migration
-
A process that occurs after strand invasion, where one strand of DNA is processively exchanged for another at Holliday junctions or D-loops, resulting in movement of the junction.
- Homologous recombination
-
(HR). The most common form of HDR, whereby exchange of genetic material occurs between two homologous chromosomes.
- ALT-associated promyelocytic leukaemia bodies
-
(APBs). Membraneless structures formed by phase separation that promote the aggregation of homologous recombination proteins, nucleases, telomere-associated proteins, PML proteins and telomeric DNA. APBs are a biomarker of alternative lengthening of telomeres (ALT).
- Small ubiquitin-like modifier
-
(SUMO). Units that are covalently attached to proteins post-translationally in a process known as sumoylation. This can alter several properties of the protein, including protein stability, localization, and addition or removal of protein–protein binding sites.
- Break-induced replication
-
Recombination-dependent DNA synthesis that initiates from a double-strand break and occurs following strand invasion mediated by RAD51 or RAD52.
- Extrachromosomal telomeric repeats
-
(ECTRs). Linear and circular extrachromosomal copies of telomeric sequences that are generated during homologous recombination in cells using ALT, including C-circles and t-circles. ECTRs are a biomarker of ALT.
- Myelofibrosis
-
A rare type of bone marrow cancer that prevents the production of blood cells, leading to anaemia and scar tissue in the bone marrow.
- Transcription elongation
-
A step in RNA transcription that occurs following initiation and prior to termination when the RNA sequence is synthesized complementary to the DNA template.
- Inhibitory concentration 50
-
(IC50). The dose of an agent required to inhibit 50% of cell growth.
- Epitope spreading
-
The process by which epitopes, distinct from the inducing epitope of a vaccine, become major targets of the immune response.
- Macrocyclic compound
-
A compound made up of chemical ring structures that each consist of 12 or more carbon atoms.
- Porphyrin
-
A molecule that consists of a ring of four linked heterocyclic groups that can be held together by a central metal atom.
- Non-homologous end-joining
-
A repair pathway where double-strand breaks are ligated together. Non-homologous end-joining (NHEJ) consists of either canonical NHEJ or alternative NHEJ. In the canonical pathway, the two ends of the DNA are bound by Ku70 and Ku80 and DNA-PKcs, which come together to form the synaptic complex. This is then ligated together by the ligase IV–XRCC4 complex. Alternative NHEJ occurs independently of canonical NHEJ proteins and involves the direct joining of short sequence homologies (microhomologies).
- Mitotic DNA synthesis
-
The process of DNA repair synthesis during mitosis.
Rights and permissions
About this article
Cite this article
Gao, J., Pickett, H.A. Targeting telomeres: advances in telomere maintenance mechanism-specific cancer therapies. Nat Rev Cancer 22, 515–532 (2022). https://doi.org/10.1038/s41568-022-00490-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-022-00490-1
This article is cited by
-
High expression of RTEL1 predicates worse progression in gliomas and promotes tumorigenesis through JNK/ELK1 cascade
BMC Cancer (2024)
-
Silencing PinX1 enhances radiosensitivity and antitumor-immunity of radiotherapy in non-small cell lung cancer
Journal of Translational Medicine (2024)
-
Drug repurposing for cancer therapy
Signal Transduction and Targeted Therapy (2024)
-
Telomere length and cancer risk: finding Goldilocks
Biogerontology (2024)
-
Giardia telomeres and telomerase
Parasitology Research (2024)