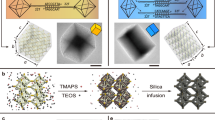
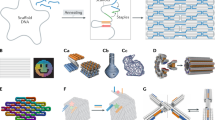
Abstract
The combination of lithographic methods with two-dimensional DNA origami self-assembly has led, among others, to the development of photonic crystal cavity arrays and the exploration of sensing nanoarrays where molecular devices are patterned on the sub-micrometre scale. Here we extend this concept to the third dimension by mounting three-dimensional DNA origami onto nanopatterned substrates, followed by silicification to provide hybrid DNA–silica structures exhibiting mechanical and chemical stability and achieving feature sizes in the sub-10-nm regime. Our versatile and scalable method relying on self-assembly at ambient temperatures offers the potential to three-dimensionally position any inorganic and organic components compatible with DNA origami nanoarchitecture, demonstrated here with gold nanoparticles. This way of nanotexturing could provide a route for the low-cost production of complex and three-dimensionally patterned surfaces and integrated devices designed on the molecular level and reaching macroscopic dimensions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in ref. 50. The data supporting the findings of this study are available within this article and its Supplementary Information.
References
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003).
Aldaye, F. A., Palmer, A. L. & Sleiman, H. F. Assembling materials with DNA as the guide. Science 321, 1795–1799 (2008).
Wang, P. et al. Magnetic plasmon networks programmed by molecular self-assembly. Adv. Mater. 31, 1901364 (2019).
Liu, X. et al. Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Kolbeck, P. J. et al. A DNA origami fiducial for accurate 3D atomic force microscopy imaging. Nano Lett. 23, 1236–1243 (2023).
Kabusure, K. M. et al. Optical characterization of DNA origami-shaped silver nanoparticles created through biotemplated lithography. Nanoscale 14, 9648–9654 (2022).
Diagne, C. T., Brun, C., Gasparutto, D., Baillin, X. & Tiron, R. DNA origami mask for sub-ten-nanometer lithography. ACS Nano 10, 6458–6463 (2016).
Surwade, S. P., Zhao, S. & Liu, H. Molecular lithography through DNA-mediated etching and masking of SiO2. J. Am. Chem. Soc. 133, 11868–11871 (2011).
Jin, Z. et al. Metallized DNA nanolithography for encoding and transferring spatial information for graphene patterning. Nat. Commun. 4, 1663 (2013).
Shen, B. et al. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 4, eaap8978 (2018).
Surwade, S. P. et al. Nanoscale growth and patterning of inorganic oxides using DNA nanostructure templates. J. Am. Chem. Soc. 135, 6778–6781 (2013).
Shen, J., Sun, W., Liu, D., Schaus, T. & Yin, P. Three-dimensional nanolithography guided by DNA modular epitaxy. Nat. Mater. 20, 683–690 (2021).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Maune, H. T. et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol. 5, 61–66 (2010).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Voigt, N. V. et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 5, 200–203 (2010).
Knudsen, J. B. et al. Routing of individual polymers in designed patterns. Nat. Nanotechnol. 10, 892–898 (2015).
Hartl, C. et al. Position accuracy of gold nanoparticles on DNA origami structures studied with small-angle X-ray scattering. Nano Lett. 18, 2609–2615 (2018).
Funke, J. J. & Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 11, 47–52 (2016).
Kershner, R. J. et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009).
Hung, A. M. et al. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 5, 121–126 (2010).
Gopinath, A., Miyazono, E., Faraon, A. & Rothemund, P. W. K. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 535, 401–405 (2016).
Gopinath, A. et al. Absolute and arbitrary orientation of single-molecule shapes. Science 371, eabd6179 (2021).
Cervantes-Salguero, K. et al. Single molecule DNA origami nanoarrays with controlled protein orientation. Biophys. Rev. 3, 031401 (2022).
Huang, D., Patel, K., Perez-Garrido, S., Marshall, J. F. & Palma, M. DNA origami nanoarrays for multivalent investigations of cancer cell spreading with nanoscale spatial resolution and single-molecule control. ACS Nano 13, 728–736 (2019).
Gopinath, A. & Rothemund, P. W. K. Optimized assembly and covalent coupling of single-molecule DNA origami nanoarrays. ACS Nano 8, 12030–12040 (2014).
Deckman, H. W. & Dunsmuir, J. H. Natural lithography. Appl. Phys. Lett. 41, 377–379 (1982).
Shetty, R. M., Brady, S. R., Rothemund, P. W. K., Hariadi, R. F. & Gopinath, A. Bench-top fabrication of single-molecule nanoarrays by DNA origami placement. ACS Nano 15, 11441–11450 (2021).
Martynenko, I. V., Ruider, V., Dass, M., Liedl, T. & Nickels, P. C. DNA origami meets bottom-up nanopatterning. ACS Nano 15, 10769–10774 (2021).
Penzo, E., Wang, R., Palma, M. & Wind, S. J. Selective placement of DNA origami on substrates patterned by nanoimprint lithography. J. Vac. Sci. Technol. B 29, 06F205 (2011).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Kim, D.-N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2011).
Suzuki, Y., Endo, M. & Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 6, 8052 (2015).
Cao, H. H. et al. Seeding the self-assembly of DNA origamis at surfaces. ACS Nano 14, 5203–5212 (2020).
Jing, X. et al. Solidifying framework nucleic acids with silica. Nat. Protoc. 14, 2416–2436 (2019).
Wen, X. et al. 3D-printed silica with nanoscale resolution. Nat. Mater. 20, 1506–1511 (2021).
Seniutinas, G. et al. Tipping solutions: emerging 3D nano-fabrication/-imaging technologies. Nanophotonics 6, 923–941 (2017).
Tan, C. et al. 2D fin field-effect transistors integrated with epitaxial high-k gate oxide. Nature 616, 66–72 (2023).
Gottlieb, S. et al. Self-assembly morphology of block copolymers in sub-10 nm topographical guiding patterns. Mol. Syst. Des. Eng. 4, 175–185 (2019).
Ouk Kim, S. et al. Epitaxial self-assembly of block copolymers on lithographically defined nanopatterned substrates. Nature 424, 411–414 (2003).
Wickham, S. F. J. et al. Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard. Nat. Commun. 11, 5768 (2020).
Zhang, T. et al. 3D DNA origami crystals. Adv. Mater. 30, 1800273 (2018).
Shaw, A., Benson, E. & Högberg, B. Purification of functionalized DNA origami nanostructures. ACS Nano 9, 4968–4975 (2015).
Stahl, E., Martin, T. G., Praetorius, F. & Dietz, H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. 53, 12735–12740 (2014).
Lin, C., Perrault, S. D., Kwak, M., Graf, F. & Shih, W. M. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 41, e40 (2012).
Martynenko, I. et al. Site-directed placement of three-dimensional DNA origami. Dryad https://doi.org/10.5061/dryad.pg4f4qrvz and Zenodo https://doi.org/10.5281/zenodo.8119098 (2023).
Acknowledgements
We thank C. Obermayer for cleanroom assistance and S. Kempter for assistance with TEM. Besides all the group members, we thank N. Vogel (FAU Erlangen-Nürnberg) for the helpful discussions. I.V.M., G.P., X.Y. and T.L. acknowledge funding from the ERC consolidator grant ‘DNA Funs’ (Project ID: 818635). E.E., V.R. and T.L. further acknowledge support from the cluster of excellence e-conversion EXC 2089/1-390776260. This work was funded by the Federal Ministry of Education and Research (BMBF) and the Free State of Bavaria under the Excellence Strategy of the Federal Government and the Länder through the ONE MUNICH Projects Munich Multiscale Biofabrication and Enabling Quantum Communication and Imaging Applications.
Author information
Authors and Affiliations
Contributions
T.L. and I.V.M. designed this study; they also designed the DNA origami samples as well as designed and optimized the interfaces. I.V.M., E.E. and V.R. assembled and purified the DNA origami samples. G.P. and X.Y. designed, assembled and purified the DNA origami tetrapods and 24HBs as well as designed the tetrapod–AuNPs and tetrapod–24HB interfaces. I.V.M., E.E. and V.R. performed the placement experiments, surface annealing experiments, AFM and SEM measurements and data analysis with assistance from M.D. and P.A. I.V.M. and T.L. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Nayan Agarwal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–15, Figs. 1–44, Tables 1 and 2 and References.
Supplementary Table 1
DNA sequences in an Excel file.
Supplementary Data 1
The caDNAno origami designs in JSON files.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martynenko, I.V., Erber, E., Ruider, V. et al. Site-directed placement of three-dimensional DNA origami. Nat. Nanotechnol. 18, 1456–1462 (2023). https://doi.org/10.1038/s41565-023-01487-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-023-01487-z