Abstract
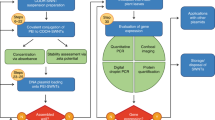
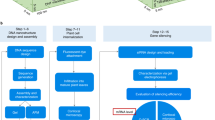
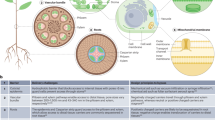
Genetic engineering of plants is at the core of sustainability efforts, natural product synthesis and crop engineering. The plant cell wall is a barrier that limits the ease and throughput of exogenous biomolecule delivery to plants. Current delivery methods either suffer from host-range limitations, low transformation efficiencies, tissue damage or unavoidable DNA integration into the host genome. Here, we demonstrate efficient diffusion-based biomolecule delivery into intact plants of several species with pristine and chemically functionalized high aspect ratio nanomaterials. Efficient DNA delivery and strong protein expression without transgene integration is accomplished in Nicotiana benthamiana (Nb), Eruca sativa (arugula), Triticum aestivum (wheat) and Gossypium hirsutum (cotton) leaves and arugula protoplasts. We find that nanomaterials not only facilitate biomolecule transport into plant cells but also protect polynucleotides from nuclease degradation. Our work provides a tool for species-independent and passive delivery of genetic material, without transgene integration, into plant cells for diverse biotechnology applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
Daniell, H., Datta, R., Varma, S., Gray, S. & Lee, S.-B. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 16, 345–348 (1998).
Liu, Y. et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 33, 301–305 (2015).
Li, T., Liu, B., Spalding, M. H., Weeks, D. P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Zhang, G. et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796 (2009).
Chen, Q. & Lai, H. Gene delivery into plant cells for recombinant protein production. Biomed. Res. Int. 2015, 932161 (2015).
Himmel, M. E. et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804 (2007).
Tufekcioglu, A., Raich, J., Isenhart, T. & Schultz, R. Biomass, carbon and nitrogen dynamics of multi-species riparian buffers within an agricultural watershed in Iowa, USA. Agroforest. Syst. 57, 187–198 (2003).
Altpeter, F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016).
Herrera-Estrella, L., Depicker, A., Van Montagu, M. & Schell, J. Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature 303, 209–213 (1983).
Baltes, N. J., Gil-Humanes, J. & Voytas, D. F. Genome engineering and agriculture: opportunities and challenges. Prog. Mol. Biol. Transl. Sci. 149, 1–26 (2017).
Klein, T. M., Wolf, E., Wu, R. & Sanford, J. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327, 70–73 (1987).
Caranta, C., Aranda M. A., Tepfer, M. & Lopez-Moya, J. J. Recent Advances in Plant Virology (Horizon Scientific, Poole, 2011).
Gleba, Y., Klimyuk, V. & Marillonnet, S. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 18, 134–141 (2007).
Song, S., Hao, Y., Yang, X., Patra, P. & Chen, J. Using gold nanoparticles as delivery vehicles for targeted delivery of chemotherapy drug fludarabine phosphate to treat hematological cancers. J. Nanosci. Nanotechnol. 16, 2582–2586 (2016).
Mizrachi, A. et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat. Commun. 8, 14292 (2017).
Demirer, G. S. & Landry, M. P. Delivering genes to plants. Chem. Eng. Progr. 113, 40–45 (2017).
Hussain, H. I., Yi, Z., Rookes, J. E., Kong, L. X. & Cahill, D. M. Mesoporous silica nanoparticles as a biomolecule delivery vehicle in plants. J. Nanoparticle Res. 15, 1676 (2013).
Silva, A. T., Nguyen, A., Ye, C., Verchot, J. & Moon, J. H. Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts. BMC Plant Biol. 10, 291 (2010).
Martin-Ortigosa, S. et al. Parameters affecting the efficient delivery of mesoporous silica nanoparticle materials and gold nanorods into plant tissues by the biolistic method. Small 8, 413–422 (2012).
Liu, J. et al. Preparation of fluorescence starch-nanoparticle and its application as plant transgenic vehicle. J. Cent. South Univ. Technol. 15, 768–773 (2008).
Chang, F.-P. et al. A simple plant gene delivery system using mesoporous silica nanoparticles as carriers. J. Mater. Chem. B 1, 5279–5287 (2013).
Zhang, H. et al. DNA nanostructures coordinate gene silencing in mature plants. https://doi.org/10.1101/538678 (2019).
Asad, S. & Arshad, M. in Properties and Applications of Silicon Carbide (ed. Gerhardt, R.) Ch. 15 (InTech, London, 2011).
Mitter, N. et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 3, 16207 (2017).
Bao, W., Wan, Y. & Baluška, F. Nanosheets for delivery of biomolecules into plant cells. Trends Plant Sci. 22, 445–447 (2017).
Bao, W., Wang, J., Wang, Q., O’Hare, D. & Wan, Y. Layered double hydroxide nanotransporter for molecule delivery to intact plant cells. Sci. Rep. 6, 26738 (2016).
Wong, M. H. et al. Lipid exchange envelope penetration (LEEP) of nanoparticles for plant engineering: a universal localization mechanism. Nano Lett. 16, 1161–1172 (2016).
Giraldo, J. P. et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat. Mater. 13, 400–408 (2014).
Wu, Y., Phillips, J. A., Liu, H., Yang, R. & Tan, W. Carbon nanotubes protect DNA strands during cellular delivery. ACS Nano 2, 2023–2028 (2008).
Zheng, M. et al. DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater. 2, 338–342 (2003).
Wang, H. et al. High-yield sorting of small-diameter carbon nanotubes for solar cells and transistors. ACS Nano 8, 2609–2617 (2014).
Liu, Q. et al. Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 9, 1007–1010 (2009).
Serag, M. F. et al. Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano 5, 493–499 (2010).
Wong, M. H. et al. Nitroaromatic detection and infrared communication from wild-type plants using plant nanobionics. Nat. Mater. 16, 264–272 (2017).
Choi, J. H. & Strano, M. S. Solvatochromism in single-walled carbon nanotubes. Appl. Phys. Lett. 90, 223114 (2007).
Tinland, B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1, 178–184 (1996).
McDermott, G. P. et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal. Chem. 85, 11619–11627 (2013).
Collier, R. et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 90, 1014–1025 (2017).
Miyaoka, Y., Mayerl, S. J., Chan, A. H. & Conklin, B. R. Detection and quantification of HDR and NHEJ induced by genome editing at endogenous gene loci using droplet digital PCR. Methods Mol. Biol. 1768, 349–362 (2018).
Glowacka, K. et al. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 39, 908–917 (2016).
Dobnik, D., Stebih, D., Blejec, A., Morisset, D. & Zel, J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci. Rep. 6, 35451 (2016).
Yoshioka, H. et al. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718 (2003).
Van Kooten, O. & Snel, J. F. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25, 147–150 (1990).
Lew, T. T. S. et al. Rational design principles for the transport and subcellular distribution of nanomaterials into plant protoplasts. Small 14, e1802086 (2018).
Schaumberg, K. A. et al. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat. Methods 13, 94 (2016).
Sullivan, A. M. et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 8, 2015–2030 (2014).
Lau, W. & Sattely, E. S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349, 1224–1228 (2015).
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L. & Landry, M. P. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018).
Del Bonis-O’Donnell, J. T. et al. Engineering molecular recognition with bio-mimetic polymers on single walled carbon nanotubes. J. Visual. Exp. 119, e55030 (2017).
Yoo, S.-D., Cho, Y.-H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565 (2007).
Yang, M., Gao, Y., Li, H. & Adronov, A. Functionalization of multiwalled carbon nanotubes with polyamide 6 by anionic ring-opening polymerization. Carbon 45, 2327–2333 (2007).
Beyene, A. G., Demirer, G. S. & Landry, M. P. Nanoparticle-templated molecular recognition platforms for detection of biological analytes. Curr. Protoc. Chem. Biol. 8, 197–223 (2016).
Ma, L. et al. Enhanced Li–S batteries using amine-functionalized carbon nanotubes in the cathode. ACS Nano 10, 1050–1059 (2015).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101 (2008).
Acknowledgements
The authors acknowledge support from a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a Stanley Fahn PDF Junior Faculty Grant under award no. PF-JFA-1760, a Beckman Foundation Young Investigator Award, a USDA AFRI award, a grant from the Gordon and Betty Moore Foundation, a USDA NIFA award, support from the Chan-Zuckerberg foundation and an FFAR New Innovator Award (to M.P.L). G.S.D. is supported by a Schlumberger Foundation Faculty for the Future Fellowship. L.C. is supported by National Defense Science and Engineering Graduate (NDSEG) Fellowship and by the LAM Foundation. The authors thank C. Gee for assisting with the Imaging-PAM Maxi fluorimeter, A. Schultink and A. Ortega for helpful discussions and C. Jakobson and D. Tullman-Ercek for generously sharing their laboratory resources. The authors also acknowledge support from UC Berkeley Molecular Imaging Center (supported by the Gordon and Betty Moore Foundation), the UC Berkeley Biological Imaging Facility (supported in part by the National Institutes of Health S10 program under award no. 1S10OD018136-01), the QB3 Shared Stem Cell Facility, the Innovative Genomics Institute (IGI), and R. Zalpuri at the Electron Microscopy Lab at UC Berkeley for TEM sample preparation and imaging.
Author information
Authors and Affiliations
Contributions
G.S.D. and M.P.L. conceived of the project, designed the study and wrote the manuscript. G.S.D. performed the majority of experiments and all data analysis. H.Z. and L.C. performed AFM imaging, and H.Z. also performed nanoparticle internalization experiments into mature leaves and western blot experiments. J.L.M. performed Agrobacterium and wheat transformation experiments. N.S.G. helped with designing ddPCR experiments and performed CNT leaf toxicity confocal imaging and TIRF experiments. F.C. performed nanoparticle internalization experiments into isolated protoplasts. Y.S. performed TEM imaging of leaves. A.J.A. and R.C. prepared the plasmids used in the studies. M.-J.C. performed particle bombardment experiments. All authors have edited and commented on the manuscript, and have given their approval of the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Nanotechnology thanks Neena Mitter, Eleni Stavrinidou and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text and Supplementary Figures 1–11
Rights and permissions
About this article
Cite this article
Demirer, G.S., Zhang, H., Matos, J.L. et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 14, 456–464 (2019). https://doi.org/10.1038/s41565-019-0382-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-019-0382-5
This article is cited by
-
Non-transgenic, PAMAM co-delivery DNA of interactive proteins NbCRVP and NbCalB endows Nicotiana benthamiana with a stronger antiviral effect to RNA viruses
Journal of Nanobiotechnology (2024)
-
Construction of peptide/plasmid DNA complexes for plant gene transfection via the basic leucine zipper domain
Polymer Journal (2024)
-
Mechanism and molecular basis of apomixis in angiosperms
Vegetos (2024)
-
Application of nanotechnology and proteomic tools in crop development towards sustainable agriculture
Journal of Crop Science and Biotechnology (2024)
-
Nanoparticles in plant resistance against bacterial pathogens: current status and future prospects
Molecular Biology Reports (2024)