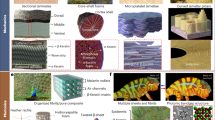
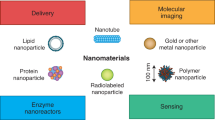
Abstract
Studying the interactions between nanoengineered materials and biological systems plays a vital role in the development of biological applications of nanotechnology and the improvement of our fundamental understanding of the bio–nano interface. A significant barrier to progress in this multidisciplinary area is the variability of published literature with regards to characterizations performed and experimental details reported. Here, we suggest a ‘minimum information standard’ for experimental literature investigating bio–nano interactions. This standard consists of specific components to be reported, divided into three categories: material characterization, biological characterization and details of experimental protocols. Our intention is for these proposed standards to improve reproducibility, increase quantitative comparisons of bio–nano materials, and facilitate meta analyses and in silico modelling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Cui, J., Richardson, J. J., Björnmalm, M., Faria, M. & Caruso, F. Nanoengineered templated polymer particles: navigating the biological realm. Acc. Chem. Res. 49, 1139–1148 (2016).
Pelaz, B. et al. Diverse applications of nanomedicine. ACS Nano 11, 2313–2381 (2017).
Malysheva, A., Lombi, E. & Voelcker, N. H. Bridging the divide between human and environmental nanotoxicology. Nat. Nanotech. 10, 835–844 (2015).
Nel, A. E. et al. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 8, 543–557 (2009).
Björnmalm, M., Faria, M. & Caruso, F. Increasing the impact of materials in and beyond bio-nano science. J. Am. Chem. Soc. 138, 13449–13456 (2016).
Mulvaney, P., Parak, W. J., Caruso, F. & Weiss, P. S. Standardizing nanomaterials. ACS Nano 10, 9763–9764 (2016).
Murashov, V. & Howard, J. (eds) Nanotechnology Standards (Springer Science & Business Media, Berlin, Heidelberg, 2011).
Pettitt, M. E. & Lead, J. R. Minimum physicochemical characterization requirements for nanomaterial regulation. Environ. Int. 52, 41–50 (2013).
Mills, K. C., Murry, D., Guzan, K. A. & Ostraat, M. L. Nanomaterial registry: database that captures the minimal information about nanomaterial physico-chemical characteristics. J. Nanoparticle Res. 16, 2219 (2014).
Boyes, W. K. et al. A comprehensive framework for evaluating the environmental health and safety implications of engineered nanomaterials. Crit. Rev. Toxicol. 47, 771–814 (2017).
Hristozov, D. et al. Frameworks and tools for risk assessment of manufactured nanomaterials. Environ. Int. 95, 36–53 (2016).
Join the dialogue. Nat. Nanotech. 7, 545 (2012).
The dialogue continues. Nat. Nanotech. 8, 69 (2013).
Schrurs, F. & Lison, D. Focusing the research efforts. Nat. Nanotech. 7, 546–548 (2012).
McCall, M. J. et al. A tiered approach. Nat. Nanotech. 8, 307–308 (2013).
Checklists work to improve science. Nature 556, 273–274 (2018).
Han, S. et al. A checklist is associated with increased quality of reporting preclinical biomedical research: a systematic review. PLoS ONE 12, e0183591 (2017).
Novère, N. L. et al. Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol. 23, 1509–1515 (2005).
Brazma, A. et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29, 365–71 (2001).
Taylor, C. F. et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 26, 889–896 (2008).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Wilhelm, S. et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016).
Bobo, D., Robinson, K. J., Islam, J., Thurecht, K. J. & Corrie, S. R. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res. 33, 2373–2387 (2016).
Kilkenny, C., Browne, W. J., Cuthill, I. C., Emerson, M. & Altman, D. G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 (2010).
Gaheen, S. et al. caNanoLab: data sharing to expedite the use of nanotechnology in biomedicine. Comput. Sci. Disc. 6, 014010 (2013).
Thomas, D. G. et al. ISA-TAB-Nano: a specification for sharing nanomaterial research data in spreadsheet-based format. BMC Biotechnol. 13, 2 (2013).
Hühn, J. et al. Selected standard protocols for the synthesis, phase transfer, and characterization of inorganic colloidal nanoparticles. Chem. Mater. 29, 399–461 (2017).
Dawidczyk, C. M., Russell, L. M. & Searson, P. C. Recommendations for benchmarking preclinical studies of nanomedicines. Cancer Res. 75, 4016–4020 (2015).
Considering Whether an FDA-Regulated Product Involves the Application of Nanotechnology (FDA, 2018); https://www.fda.gov/regulatoryinformation/guidances/ucm257698.htm
Albanese, A., Tang, P. S. & Chan, W. C. W. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 14, 1–16 (2012).
Buriak, J. M. Preface to the special issue on methods and protocols in materials chemistry. Chem. Mater. 29, 1–2 (2017).
Behzadi, S. et al. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 46, 4218–4244 (2017).
Hadjidemetriou, M. & Kostarelos, K. Nanomedicine: Evolution of the nanoparticle corona. Nat. Nanotech 12, 288–290 (2017).
Satzer, P., Svec, F., Sekot, G. & Jungbauer, A. Protein adsorption onto nanoparticles induces conformational changes: particle size dependency, kinetics, and mechanisms. Eng. Life Sci. 16, 238–246 (2016).
Talamini, L. et al. Influence of size and shape on the anatomical distribution of endotoxin-free gold nanoparticles. ACS Nano 11, 5519–5529 (2017).
Soo Choi, H. et al. Renal clearance of quantum dots. Nat. Biotechnol. 25, 1165–1170 (2007).
Jiang, W., Kim, B. Y. S., Rutka, J. T. & Chan, W. C. W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotech. 3, 145–150 (2008).
Walkey, C. D., Olsen, J. B., Guo, H., Emili, A. & Chan, W. C. W. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 134, 2139–2147 (2012).
Sperling, R. A. et al. Size determination of (bio)conjugated water-soluble colloidal nanoparticles: a comparison of different techniques. J. Phys. Chem. C 111, 11552–11559 (2007).
Moore, T. L. et al. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 44, 6287–6305 (2015).
Glass, J. J. et al. Charge has a marked influence on hyperbranched polymer nanoparticle association in whole human blood. ACS Macro Lett. 6, 586–592 (2017).
Walkey, C. D. et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 8, 2439–2455 (2014).
Doane, T. L., Chuang, C.-H., Hill, R. J. & Burda, C. Nanoparticle ζ-potentials. Acc. Chem. Res. 45, 317–326 (2012).
Hinderliter, P. M. et al. ISDD: A computational model of particle sedimentation, diffusion and target cell dosimetry for in vitro toxicity studies. Part. Fibre Toxicol 7, 36 (2010).
Cui, J. et al. A framework to account for sedimentation and diffusion in particle–cell interactions. Langmuir 32, 12394–12402 (2016).
DeLoid, G. M., Cohen, J. M., Pyrgiotakis, G. & Demokritou, P. Preparation, characterization, and in vitro dosimetry of dispersed, engineered nanomaterials. Nat. Protoc. 12, 355–371 (2017).
Mura, S., Nicolas, J. & Couvreur, P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 12, 991–1003 (2013).
Allen, T. M. Drug delivery systems: entering the mainstream. Science 303, 1818–1822 (2004).
Howard, C. B. et al. Overcoming instability of antibody-nanomaterial conjugates: next generation targeted nanomedicines using bispecific antibodies. Adv. Healthcare Mater. 5, 2055–2068 (2016).
Kamphuis, M. M. J. et al. Targeting of cancer cells using click-functionalized polymer capsules. J. Am. Chem. Soc. 132, 15881–15883 (2010).
Ju, Y. et al. Engineered metal-phenolic capsules show tunable targeted delivery to cancer cells. Biomacromolecules 17, 2268–2276 (2016).
Colombo, M. et al. Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies. Nat. Commun. 7, 13818 (2016).
Torrice, M. Does nanomedicine have a delivery problem? ACS Cent. Sci. 2, 434–437 (2016).
Lammers, T. et al. Cancer nanomedicine: is targeting our target? Nat. Rev. Mater. 1, 16069 (2016).
Björnmalm, M., Thurecht, K. J., Michael, M., Scott, A. M. & Caruso, F. Bridging bio–nano science and cancer nanomedicine. ACS Nano 11, 9594–9613 (2017).
Herda, L. M., Hristov, D. R., Lo Giudice, M. C., Polo, E. & Dawson, K. A. Mapping of molecular structure of the nanoscale surface in bionanoparticles. J. Am. Chem. Soc. 139, 111–114 (2017).
Peng, H.-S. & Chiu, D. T. Soft fluorescent nanomaterials for biological and biomedical imaging. Chem. Soc. Rev. 44, 4699–4722 (2015).
Puttick, S., Boase, N. R. B., Blakey, I. & Thurecht, K. J. Imaging tumour distribution of a polymeric drug delivery platform in vivo by PET-MRI. J. Chem. Technol. Biotechnol. 90, 1237–1244 (2015).
Wang, K., Peng, H., Thurecht, K. J., Puttick, S. & Whittaker, A. K. Multifunctional hyperbranched polymers for CT/19F MRI bimodal molecular imaging. Polym. Chem. 7, 1059–1069 (2016).
Rolfe, B. E. et al. Multimodal polymer nanoparticles with combined19F magnetic resonance and optical detection for tunable, targeted, multimodal imaging in vivo. J. Am. Chem. Soc. 136, 2413–2419 (2014).
Michalet, X. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 (2005).
Schwartz, A. et al. Formalization of the MESF unit of fluorescence intensity. Cytometry 57B, 1–6 (2004).
Anselmo, A. C. & Mitragotri, S. Impact of particle elasticity on particle-based drug delivery systems. Adv. Drug Delivery Rev. 108, 51–67 (2017).
Berry, J. D., Mettu, S. & Dagastine, R. R. Precise measurements of capsule mechanical properties using indentation. Soft Matter 13, 1943–1947 (2017).
Teeguarden, J. G., Hinderliter, P. M., Orr, G., Thrall, B. D. & Pounds, J. G. Particokinetics in vitro: dosimetry considerations for in vitro nanoparticle toxicity assessments. Toxicol. Sci. 95, 300–312 (2007).
Feliu, N., Sun, X., Alvarez Puebla, R. A. & Parak, W. J. Quantitative particle–cell interaction: some basic physicochemical pitfalls. Langmuir 33, 6639–6646 (2017).
Almeida, J. L., Cole, K. D. & Plant, A. L. Standards for cell line authentication and beyond. PLoS Biol. 14, e1002476 (2016).
Olarerin-George, A. O. & Hogenesch, J. B. Assessing the prevalence of mycoplasma contamination in cell culture via a survey of NCBI’s RNA-seq archive. Nucleic Acids Res. 43, 2535–2542 (2015).
Kim, J. A., Åberg, C., Salvati, A. & Dawson, K. A. Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat. Nanotech. 7, 62–68 (2012).
Morgan, R. A. Human tumor xenografts: the good, the bad, and the ugly. Mol. Ther. 20, 882–884 (2012).
Yan, Y. et al. Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano 7, 10960–10970 (2013).
Delmaar, C. J. E. et al. A practical approach to determine dose metrics for nanomaterials. Environ. Toxicol. Chem. 34, 1015–1022 (2015).
Johnston, A. P. R. Life under the microscope: quantifying live cell interactions to improve nanoscale drug delivery. ACS Sensors 2, 4–9 (2017).
Hibbert, D. B. Quality Assurance in the Analytical Chemistry Laboratory (Oxford University Press, Oxford, 2007).
OpenDataFit. Available at http://supramolecular.org/. (Accessed 3 January 2018).
Joining the reproducibility initiative. Nat. Nanotech. 9, 949 (2014).
Scrutinizing lasers. Nat. Photon. 11, 139 (2017).
Nosek, B. A. et al. Promoting an open research culture. Science 348, 1422–1425 (2015).
Chen, R. & Riviere, J. E. Biological and environmental surface interactions of nanomaterials: characterization, modeling, and prediction. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 9, e1440 (2017).
Acknowledgements
This work was supported by the Australian Research Council (ARC) Centre of Excellence in Convergent Bio-Nano Science and Technology (Project Number CE140100036) and under the ARC Australian Laureate Fellowship scheme (F.C., FL120100030; T.P.D., FL140100052; J.J.G., FL150100060). M.B. acknowledges support from H2020/European Commission through a Marie Skłodowska-Curie Individual Fellowship under grant agreement no. 745676. F.C. acknowledges the award of a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellowship (APP1135806). M.K. receives support from NHMRC Principal Research Fellowship (M.K.; APP1119152) and Cancer Council New South Wales Program Grant (PG16-01). M.K., T.P.D. and J.J.G. receive support from NHMRC Program Grant (APP1091261). A.P.R.J. receives support from the NHMRC Career Development Fellowship (APP1141551) and NHMRC Project Grants (APP1129672, APP1124161). R.G.P. receives support from NHMRC Senior Principal Research Fellowship (APP1058565) and NHMRC Program Grant (APP1037320). W.J.P. acknowledges funding from the Deutsche Forschungsgemeinschaft (project DFG PA 794/28-1) and a visiting professor fellowship from the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology. M.M.S. acknowledges support from the ERC Seventh Framework Programme Consolidator grant “Naturale CG” [616417], the i-sense EPSRC IRC in Early Warning Sensing Systems for Infectious Diseases (EP/K031953/1) and the EPSRC grant “Bio-functionalised nanomaterials for ultrasensitive biosensing” (EP/K020641/1). The authors thank A. E. Burke and C. Lynm for assistance with the preparation of Fig. 1. The development of these guidelines was led by the ARC Centre of Excellence in Convergent Bio-Nano Science and Technology (https://www.cbns.org.au/).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3
Rights and permissions
About this article
Cite this article
Faria, M., Björnmalm, M., Thurecht, K.J. et al. Minimum information reporting in bio–nano experimental literature. Nature Nanotech 13, 777–785 (2018). https://doi.org/10.1038/s41565-018-0246-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-018-0246-4
This article is cited by
-
Subcutaneous drug delivery from nanoscale systems
Nature Reviews Bioengineering (2024)
-
Human and environmental safety of carbon nanotubes across their life cycle
Nature Reviews Materials (2023)
-
New opportunities and old challenges in the clinical translation of nanotheranostics
Nature Reviews Materials (2023)
-
Nanomaterial-based contrast agents
Nature Reviews Methods Primers (2023)
-
The protein corona from nanomedicine to environmental science
Nature Reviews Materials (2023)