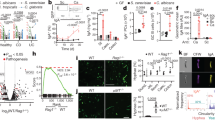
Abstract
Secretory immunoglobulin A (sIgA) plays an important role in gut barrier protection by shaping the resident microbiota community, restricting the growth of bacterial pathogens and enhancing host protective immunity via immunological exclusion. Here, we found that a portion of the microbiota-driven sIgA response is induced by and directed towards intestinal fungi. Analysis of the human gut mycobiota bound by sIgA revealed a preference for hyphae, a fungal morphotype associated with virulence. Candida albicans was a potent inducer of IgA class-switch recombination among plasma cells, via an interaction dependent on intestinal phagocytes and hyphal programming. Characterization of sIgA affinity and polyreactivity showed that hyphae-associated virulence factors were bound by these antibodies and that sIgA influenced C. albicans morphotypes in the murine gut. Furthermore, an increase in granular hyphal morphologies in patients with Crohn’s disease compared with healthy controls correlated with a decrease in antifungal sIgA antibody titre with affinity to two hyphae-associated virulence factors. Thus, in addition to its importance in gut bacterial regulation, sIgA targets the uniquely fungal phenomenon of hyphal formation. Our findings indicate that antifungal sIgA produced in the gut can play a role in regulating intestinal fungal commensalism by coating fungal morphotypes linked to virulence, thereby providing a protective mechanism that might be dysregulated in patients with Crohn’s disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data are provided with this paper. The data that support the findings of this study are available from the corresponding author on request.
References
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host–microbiota mutualism. Science 325, 617 (2009).
Cerutti, A. & Rescigno, M. The biology of intestinal immunoglobulin A responses. Immunity 28, 740–750 (2008).
Pabst, O. & Slack, E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 13, 12–21 (2020).
Spencer, J. & Sollid, L. M. The human intestinal B-cell response. Mucosal Immunol. 9, 1113–1124 (2016).
Bunker, J. J. & Bendelac, A. IgA responses to microbiota. Immunity 49, 211–224 (2018).
Bunker, J. J. et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358, eaan6619 (2017).
Bunker, J. J. et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015).
Uchimura, Y. et al. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity 49, 545–559.e545 (2018).
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011).
Macpherson, A. J. et al. IgA production without mu or delta chain expression in developing B cells. Nat. Immunol. 2, 625–631 (2001).
Gopalakrishna, K. P. et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 25, 1110–1115 (2019).
Nowosad, C. R. et al. Tunable dynamics of B cell selection in gut germinal centres. Nature 588, 321–326 (2020).
Chen, H. et al. BCR selection and affinity maturation in Peyer’s patch germinal centres. Nature 582, 421–425 (2020).
Kabbert, J. et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J. Exp. Med. 217, e20200275 (2020).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Viladomiu, M. et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci. Transl. Med. 9, eaaf9655 (2017).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2017).
Liguori, G. et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohn’s Colitis 10, 296–305 (2015).
Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500 (2015).
Hoarau, G. et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7, e01250-16 (2016).
Limon, J. J. et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388.e6 (2019).
Leonardi, I. et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe 27, 823–829.e3 (2020).
Leonardi, I. et al. CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236 (2018).
Jain, U. et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 371, 1154–1159 (2021).
Yang, A. M. et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841 (2017).
Israeli, E. et al. Anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic antibodies as predictors of inflammatory bowel disease. Gut 54, 1232–1236 (2005).
Standaert-Vitse, A. et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am. J. Gastroenterol. 104, 1745–1753 (2009).
Doron, I. et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 184, 1017–1031.e1014 (2021).
Millet, N., Solis, N. V. & Swidergall, M. Mucosal IgA prevents commensal Candida albicans dysbiosis in the oral cavity. Front. Immunol. 11, 555363 (2020).
Witchley, J. N. et al. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe 25, 432–443.e6 (2019).
Liang, S.-H. et al. Hemizygosity enables a mutational transition governing fungal virulence and commensalism. Cell Host Microbe 25, 418–431.e6 (2019).
Gow, N. A. R. & Hube, B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412 (2012).
Doron, I., Leonardi, I. & Iliev, I. D. Profound mycobiome differences between segregated mouse colonies do not influence Th17 responses to a newly introduced gut fungal commensal. Fungal Genet. Biol. 127, 45–49 (2019).
Koch, M. A. et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016).
Macpherson, A. J. et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Smith, K., McCoy, K. D. & Macpherson, A. J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69 (2007).
Senda, S., Cheng, E. & Kawanishi, H. Aging-associated changes in murine intestinal immunoglobulin A and M secretions. Scand. J. Immunol. 27, 157–164 (1988).
Lécuyer, E. et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620 (2014).
Fan, D. et al. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 21, 808–814 (2015).
Zhai, B. et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat. Med. 26, 59–64 (2020).
Li, X. et al. Response to fungal dysbiosis by gut-resident CX3CR1(+) mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe 24, 847–856.e4 (2018).
Schaedler, R. W., Dubs, R. & Costello, R. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122, 77–82 (1965).
Tso, G. H. W. et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589 (2018).
Pande, K., Chen, C. & Noble, S. M. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 45, 1088–1091 (2013).
Pierce, J. V., Dignard, D., Whiteway, M. & Kumamoto, C. A. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot. Cell 12, 37 (2013).
Pierce, J. V. & Kumamoto, C. A. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 3, e00117–12 (2012).
Allert, S. et al. Candida albicans-induced epithelial damage mediates translocation through intestinal barriers. mBio 9, e00915–18 (2018).
Lo, H. J. et al. Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939–949 (1997).
Hube, B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr. Opin. Microbiol. 7, 336–341 (2004).
Mowat, A. M. & Agace, W. W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685 (2014).
Koscsó, B. et al. Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ. Sci. Immunol. 5, eaax0062 (2020).
Farache, J., Zigmond, E., Shakhar, G. & Jung, S. Contributions of dendritic cells and macrophages to intestinal homeostasis and immune defense. Immunol. Cell Biol. 91, 232–239 (2013).
Bogunovic, M., Mortha, A., Muller, P. A. & Merad, M. Mononuclear phagocyte diversity in the intestine. Immunol. Res 54, 37–49 (2012).
Chikina, A. S. et al. Macrophages maintain epithelium integrity by limiting fungal product absorption. Cell 183, 411–428.e16 (2020).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Joeris, T., Müller-Luda, K., Agace, W. W. & Mowat, A. M. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. 10, 845–864 (2017).
Kubinak, J. L. et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163 (2015).
Macpherson, A. J., McCoy, K. D., Johansen, F. E. & Brandtzaeg, P. The immune geography of IgA induction and function. Mucosal Immunol. 1, 11–22 (2008).
Chen, K., Magri, G., Grasset, E. K. & Cerutti, A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat. Rev. Immunol. 20, 427–441 (2020).
Ha, S. A. et al. Regulation of B1 cell migration by signals through Toll-like receptors. J. Exp. Med. 203, 2541–2550 (2006).
Netea, M. G., Joosten, L. A. B., van der Meer, J. W. M., Kullberg, B.-J. & van de Veerdonk, F. L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15, 630–642 (2015).
Li, X. V., Leonardi, I. & Iliev, I. D. Gut mycobiota in immunity and inflammatory disease. Immunity 50, 1365–1379 (2019).
Liu, Y. & Filler, S. G. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10, 168–173 (2011).
Moyes, D. L. et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68 (2016).
Fransen, F. et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 43, 527–540 (2015).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Shimoda, M., Inoue, Y., Azuma, N. & Kanno, C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer’s patches. Immunology 97, 9–17 (1999).
Brand, A. Hyphal growth in human fungal pathogens and its role in virulence. Int. J. Microbiol. 2012, 517529 (2012).
Wu, G. et al. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11, e1005614 (2015).
Saadatzadeh, M. R., Ashbee, H. R., Holland, K. T. & Ingham, E. Production of the mycelial phase of Malassezia in vitro. Med. Mycol. 39, 487–493 (2001).
Loures, F. V. et al. Recognition of Aspergillus fumigatus hyphae by human plasmacytoid dendritic cells is mediated by dectin-2 and results in formation of extracellular traps. PLoS Pathog. 11, e1004643 (2015).
Moyes, D. L. et al. Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PLoS ONE 6, e26580 (2011).
Zuza-Alves, D. L., Silva-Rocha, W. P. & Chaves, G. M. An update on Candida tropicalis based on basic and clinical approaches. Front. Microbiol. 8, 1927–1927 (2017).
Gantner, B. N., Simmons, R. M. & Underhill, D. M. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24, 1277–1286 (2005).
Lin, X., Alspaugh, J. A., Liu, H. & Harris, S. Fungal morphogenesis. Cold Spring Harb. Perspect. Med. 5, a019679 (2014).
McKenzie, C. G. et al. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78, 1650–1658 (2010).
Ost, K. S. et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118 (2021).
Staab, J. F. & Sundstrom, P. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14, 681–686 (1998).
Fonzi, W. A. & Irwin, M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728 (1993).
Prieto, D., Román, E., Correia, I. & Pla, J. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9, e87128 (2014).
Park, Y. N. & Morschhäuser, J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4, 1328–1342 (2005).
Noble, S. M. & Johnson, A. D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298–309 (2005).
Granger, B. L., Flenniken, M. L., Davis, D. A., Mitchell, A. P. & Cutler, J. E. Yeast wall protein 1 of Candida albicans. Microbiology (Reading) 151, 1631–1644 (2005).
Román, E., Coman, I., Prieto, D., Alonso-Monge, R. & Pla, J. Implementation of a CRISPR-based system for gene regulation in Candida albicans. mSphere 4, e00001–e00019 (2019).
Chauvel, M. et al. A versatile overexpression strategy in the pathogenic yeast Candida albicans: identification of regulators of morphogenesis and fitness. PLoS ONE 7, e45912 (2012).
Pla, J., Pérez-Díaz, R. M., Navarro-García, F., Sánchez, M. & Nombela, C. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene 165, 115–120 (1995).
Xie, J. et al. White–opaque switching in natural MTLa/α isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 11, e1001525 (2013).
Acknowledgements
We thank members of the Iliev laboratory for their critical reviews of the manuscript. We thank Ramnik Xavier for discussion and analysis that helped us with shaping the hypothesis. We thank all contributing members of the JRI IBD Live Cell Bank Consortium, and the Microbiome Core Laboratory of Weill Cornell Medicine. Support for human sample acquisition through the JRI IBD Live Cell Bank is provided by the Jill Roberts Institute, Jill Roberts Center for IBD, Cure for IBD, the Rosanne H. Silbermann Foundation and Weill Cornell Medicine Division of Pediatric Gastroenterology and Nutrition. J.P. and E.R. were funded by PGC2018-095047-B-I00 from MINECO and InGEMICS (B2017/BMD-3691) from CAM. Research in the Iliev laboratory is supported by US National Institutes of Health (R01AI163007, R01DK113136 and R01DK121977), the Leona M. and Harry B. Helmsley Charitable Trust, the Irma T. Hirschl Career Scientist Award, Crohn’s and Colitis Foundation, Pilot Project Funding from the Center for Advanced Digestive Care (CADC) and the Burrough Welcome Trust PATH Award.
Author information
Authors and Affiliations
Contributions
I.D. and I.D.I. conceived and designed the experiments. I.D., M.M., D.G.S., X.V.L., I.L., T.K., W.D.F., W.-Y.L., E.R. and M.B.-D. performed the experiments. J.P., R.S.L. and P.C.W., generated key research materials and contributed to interpretation of the experiments. I.D. and I.D.I. generated figures and legends from analysed data. I.D.I acquired funding for the project. I.D. and I.D.I. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Kathy McCoy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of gut fungi from fecal material by flow cytometry and anti-C. albicans sIgA dynamics.
a, Microbes in fecal material from SPF WT WCM-CE mice were distinguished as a Sybrhi population that is absent in GF mouse feces. b, Fungi (SybrhiCFW+) were enriched from bacteria (SybrhiCFW−) through size separation by 900g centrifugation and calcofluor white (CFW) staining of the resulting pellet. c, C. albicans cultured for 18 hours in hyphae-inductive media was stained with fecal supernatant from C. albicans-colonized GF mice (N = 6) collected at 0, 2-, 4−, 8- and 14-days post colonization, followed by sIgA staining. Analysis of IgA binding representative of two independent experiments, one-way ANOVA, followed by Sidak’s test. d, Representative flow cytometry plots of frequency of B220+IgA+ among Live CD45+CD4− cells in the PP of germ-free (GF) mice orally gavaged with PBS (GF) or colonized for two weeks with C. albicans (+Ca). Data in (c) represents mean ± SEM.
Extended Data Fig. 2 CFW+Sybrhi FSChiSSChi C. albicans population in feces represents hyphal/ pseudohyphal fungal morphologies that are preferentially bound by sIgA.
a, CFW+Sybrhi fungal population from feces of SPF mice colonized with CAF2-RFP C. albicans was sorted into FSChiSSChi and FSCl°SSClo fractions. Constitutive expression of RFP in this strain allows for high visibility and resistance to signal quenching upon prolonged light exposure during flow cytometry and microscopy on the same material. b, Immunofluorescence microscopy of sorted material from (a). Composite images at 20X magnification of FSChiSSChi and FSCloSSClo shown in left and right panels, respectively. Scale bar represents 25μm. Data representative of two independent experiments. c, CFW+Sybrhi fungal population from feces of SPF mice colonized with CAF2-RFP was sorted into IgA+ and IgA− populations. Gray histograms represent IgA−isotype control staining used to distinguish sorted populations. d-e, Area (d) and perimeter length (e) of CAF2-RFP were compared between IgA+ and IgA− sorted populations. Data represents two independent experiments, mean ± SEM. Two-sided Mann-Whitney test. N = 5.
Extended Data Fig. 3 Assessment of Ca-dREP C. albicans double reporter strain upon IgA staining and hyphae forming deficiency of efg1Δ/Δ cph1Δ/Δ C. albicans strain.
a-b, Immunofluorescence microscopy of Ca-dREP incubated with human fecal supernatant as a source of sIgA and stained with DAPI and anti-human IgA−APC (a) or an APC isotype control (b). Single channel staining of 2 samples shown. Left to right: DAPI, constitutive ENO1-GFP expression, hyphae-specific HWP1-RFP expression, and anti-human IgA−APC (a) or APC isotype control (b). Top rows in a and b correspond to composite images in Fig. 2d,e, representing three independent experiments. Scale bar represents 50μm. c, Hyphae-competent (WT), but not hyphae-deficient (yeast-locked; efg1Δ/Δ cph1Δ/Δ) strains of C. albicans forms hyphae upon hyphae-inducing stimuli in vitro. Scale bar represents 25μm.
Extended Data Fig. 4 Flow cytometry gating strategy in PPs, LP and in feces.
a-b, Cell gating startegy for assessment of IgA+ GC B cell in PPs (a) and IgA+ plasmablasts in lamina propria (b). c, gating strategy of C.albicans cells in feces pre- and post- C.albicans (C.a) colonization.
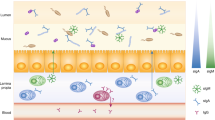
Extended Data Fig. 5 Graphical abstract for the model of antifungal IgA induction by and regulation of intestinal fungal commensalism.
(Credit: Created with BioRender.com).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2. Supplementary Table 1. Reagent sources. Supplementary Table 2. Mucosal washings and serum metadata.
Source data
Source Data Fig. 1
Flow cytometry and ELISA data.
Source Data Fig. 2
Flow cytometry, microscopy and microbial counts data.
Source Data Fig. 3
Flow cytometry, microbial counts and ELISA data.
Source Data Fig. 4
Flow cytometry data.
Source Data Fig. 5
Flow cytometry and ELISA data.
Source Data Extended Data Fig. 1
Flow cytometry data.
Source Data Extended Data Fig. 2
Flow cytometry and microscopy data.
Rights and permissions
About this article
Cite this article
Doron, I., Mesko, M., Li, X.V. et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nat Microbiol 6, 1493–1504 (2021). https://doi.org/10.1038/s41564-021-00983-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-00983-z
This article is cited by
-
The hyphal-specific toxin candidalysin promotes fungal gut commensalism
Nature (2024)
-
Veränderte T-Zell-Reaktion gegen Hefepilze bei chronischer Darmentzündung
BIOspektrum (2024)
-
A comprehensive guide to assess gut mycobiome and its role in pathogenesis and treatment of inflammatory bowel disease
Indian Journal of Gastroenterology (2024)
-
Secretory IgA reduced the ergosterol contents of Candida albicans to repress its hyphal growth and virulence
Applied Microbiology and Biotechnology (2024)
-
Improved eukaryotic detection compatible with large-scale automated analysis of metagenomes
Microbiome (2023)