Abstract
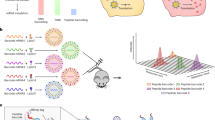
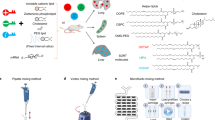
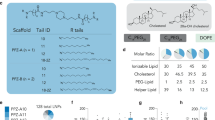
To unlock the full promise of messenger (mRNA) therapies, expanding the toolkit of lipid nanoparticles is paramount. However, a pivotal component of lipid nanoparticle development that remains a bottleneck is identifying new ionizable lipids. Here we describe an accelerated approach to discovering effective ionizable lipids for mRNA delivery that combines machine learning with advanced combinatorial chemistry tools. Starting from a simple four-component reaction platform, we create a chemically diverse library of 584 ionizable lipids. We screen the mRNA transfection potencies of lipid nanoparticles containing those lipids and use the data as a foundational dataset for training various machine learning models. We choose the best-performing model to probe an expansive virtual library of 40,000 lipids, synthesizing and experimentally evaluating the top 16 lipids flagged. We identify lipid 119-23, which outperforms established benchmark lipids in transfecting muscle and immune cells in several tissues. This approach facilitates the creation and evaluation of versatile ionizable lipid libraries, advancing the formulation of lipid nanoparticles for precise mRNA delivery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information files.
Code availability
The source code used for the ML algorithm is available in Supplementary Information.
References
Miao, L., Zhang, Y. & Huang, L. mRNA vaccine for cancer immunotherapy. Mol. Cancer 20, 41 (2021).
Tartof, S. Y. et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet 398, 1407–1416 (2021).
Trepotec, Z., Lichtenegger, E., Plank, C., Aneja, M. K. & Rudolph, C. Delivery of mRNA therapeutics for the treatment of hepatic diseases. Mol. Ther. 27, 794–802 (2019).
Blanchard, E. L. et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 39, 717–726 (2021).
Qiu, M. et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl Acad. Sci. USA 118, e2020401118 (2021).
Neklesa, T. K., Winkler, J. D. & Crews, C. M. Targeted protein degradation by PROTACs. Pharm. Ther. 174, 138–144 (2017).
Kim, M. et al. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv. 7, eabf4398 (2021).
Han, X. et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 12, 7233 (2021).
Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28, 172–176 (2010).
Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Zhang, Y., Sun, C., Wang, C., Jankovic, K. E. & Dong, Y. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021).
Qiu, M. et al. Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl Acad. Sci. USA 119, e2116271119 (2022).
Li, B. et al. Enhancing the immunogenicity of lipid-nanoparticle mRNA vaccines by adjuvanting the ionizable lipid and the mRNA. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-023-01082-6 (2023).
Li, B. et al. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat. Biotechnol. 41, 1410–1415 (2023).
Akinc, A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 (2008).
Whitehead, K. A. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 5, 4277 (2014).
Dong, Y. et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl Acad. Sci. USA 111, 3955–3960 (2014).
Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019).
Chen, J. et al. Combinatorial design of ionizable lipid nanoparticles for muscle-selective mRNA delivery with minimized off-target effects. Proc. Natl Acad. Sci. USA 120, e2309472120 (2023).
Maier, M. A. et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 21, 1570–1578 (2013).
Kaczmarek, J. C. et al. Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 18, 6449–6454 (2018).
Ekins, S. et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019).
Yamankurt, G. et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat. Biomed. Eng. 3, 318–327 (2019).
Blagus, R. & Lusa, L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinform. 14, 106 (2013).
Yap, C. W. PaDEL-Descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474 (2011).
Bergstra, J. & Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 13, 281–305 (2012).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Liu, S. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 20, 701–710 (2021).
Sahu, I., Haque, A. K. M. A., Weidensee, B., Weinmann, P. & Kormann, M. S. D. Recent developments in mRNA-based protein supplementation therapy to target lung diseases. Mol. Ther. J. Am. Soc. Gene Ther. 27, 803–823 (2019).
Chakraborty, C., Sharma, A. R., Bhattacharya, M. & Lee, S.-S. From COVID-19 to cancer mRNA vaccines: moving from bench to clinic in the vaccine landscape. Front. Immunol. 12, 679344 (2021).
Gan, Z. et al. Nanoparticles containing constrained phospholipids deliver mRNA to liver immune cells in vivo without targeting ligands. Bioeng. Transl. Med. 5, e10161 (2020).
Acknowledgements
This work was supported by the National Institutes of Health (grant no. R61AI161805), Translate Bio (Lexington, MA), Canadian Institutes of Health Research (nos PJH-185722 and PJT-190109) and the University of Toronto Data Science Institute Catalyst Grant. B.L. was also supported by the Leslie Dan Faculty of Pharmacy start-up fund, the Connaught Fund (no. 514681), the J. P. Bickell Foundation (grant no. 515159), the Canada Research Chairs Program (no. CRC-2022-00575), Natural Sciences and Engineering Research Council of Canada (no. RGPIN-2023-05124), the Canada Foundation for Innovation, John R. Evans Leaders Fund (no. 43711) and the Cystic Fibrosis Foundation. We acknowledge the use of resources at the Animal Imaging & Preclinical Testing and Flow Cytometry Core Facilities (Swanson Biotechnology Center, David H. Koch Institute for Integrative Cancer Research at the Massachusetts Institute of Technology).
Author information
Authors and Affiliations
Contributions
B.L. conceived the study and designed the experiments. B.L., I.O.R., A.G.R.G., L.S., T.M.R., F.A.O., A.Y.J. and A.V. performed the experiments and data analysis. B.L. and L.S. conducted the ML training, optimization and analysis. B.L., I.O.R., A.G.R.G., A.V., R.S.L. and D.G.A. wrote and edited the paper. B.L., R.S.L. and D.G.A. acquired funding and supervised the project. All authors provided feedback and helped shape the research, data analysis and paper.
Corresponding authors
Ethics declarations
Competing interests
B.L., I.O.R., A.G.R.G. and D.G.A. (inventors) have filed a patent through MIT (applicant) for the development of the described lipids (Application No.: PCT/US2023/010027). Patent status: pending. D.G.A. receives research funding from Sanofi/Translate and is a founder of ORNA Therapeutics. R.S.L. is a cofounder of Moderna Therapeutics and has been involved with a number of other entities, compensated or uncompensated; a complete list is available at https://www.dropbox.com/s/yc3xqb5s8s94v7x/Rev%20Langer%20COI.pdf?dl=0. The other authors declare no competing interests.
Peer review
Peer review information
Nature Materials thanks Roy van der Meel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Methods and source code.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, B., Raji, I.O., Gordon, A.G.R. et al. Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry. Nat. Mater. (2024). https://doi.org/10.1038/s41563-024-01867-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41563-024-01867-3