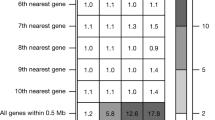
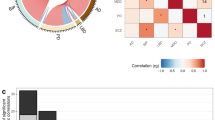
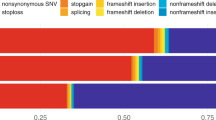
Abstract
While numerous genomic loci have been identified for neuropsychiatric conditions, the contribution of protein-coding variants has yet to be determined. Here we conducted a large-scale whole-exome-sequencing study to interrogate the impact of protein-coding variants on 46 neuropsychiatric diseases and 23 traits in 350,770 adults from the UK Biobank. Twenty new genes were associated with neuropsychiatric diseases through coding variants, among which 16 genes had impacts on the longitudinal risks of diseases. Thirty new genes were associated with neuropsychiatric traits, with SYNGAP1 showing pleiotropic effects across cognitive function domains. Pairwise estimation of genetic correlations at the coding-variant level highlighted shared genetic associations among pairs of neurodegenerative diseases and mental disorders. Lastly, a comprehensive multi-omics analysis suggested that alterations in brain structures, blood proteins and inflammation potentially contribute to the gene–phenotype linkages. Overall, our findings characterized a compendium of protein-coding variants for future research on the biology and therapeutics of neuropsychiatric phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data used in this study were accessed from the UKB (https://biobank.ndph.ox.ac.uk/) under application number 19542. Summary results of the main analyses have been made accessible through https://doi.org/10.6084/m9.figshare.25342486.v1 (ref. 70) and https://doi.org/10.6084/m9.figshare.25342201.v1 (ref. 71). The snRNA-seq data were obtained from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE173731.
Code availability
All software and R packages used to perform the analyses in this work are freely available online: SAIGE-GENE+ v.1.1.6.2, https://github.com/saigegit/SAIGE; PLINK v.2.0, https://www.cog-genomics.org/plink/2.0/; SnpEff v.5.l, https://pcingola.github.io/SnpEff/se_introduction/; ANNOVAR, https://annovar.openbioinformatics.org/en/latest/; BHR v.0.1.0, https://github.com/ajaynadig/bhr; LDSC v.1.0.1, https://github.com/bulik/ldsc/; FUMA platform, http://fuma.ctglab.nl/; SynGO, https://www.syngoportal.org/. The scripts used to conduct the main analyses are available at https://github.com/wubsfudaner/neuropsychiatric-diseases-and-traits-wes.
References
Heinzen, E. L., Neale, B. M., Traynelis, S. F., Allen, A. S. & Goldstein, D. B. The genetics of neuropsychiatric diseases: looking in and beyond the exome. Annu. Rev. Neurosci. 38, 47–68 (2015).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Ashley, E. A. Towards precision medicine. Nat. Rev. Genet. 17, 507–522 (2016).
de la Torre-Ubieta, L., Won, H., Stein, J. L. & Geschwind, D. H. Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361 (2016).
Sullivan, P. F. & Geschwind, D. H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 177, 162–183 (2019).
McCoy, T. H. Jr., Hart, K., Pellegrini, A. & Perlis, R. H. Genome-wide association identifies a novel locus for delirium risk. Neurobiol. Aging 68, 160.e169–160.e114 (2018).
Malik, R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 50, 524–537 (2018).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Power, R. A. et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry 70, 22–30 (2013).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Van Hout, C. V. et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020).
Holstege, H. et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat. Genet. 54, 1786–1794 (2022).
Kucukali, F. et al. Whole-exome rare-variant analysis of Alzheimer’s disease and related biomarker traits. Alzheimer’s Dement. 19, 2317–2331 (2022).
Auer, P. L. et al. Rare and coding region genetic variants associated with risk of ischemic stroke: the NHLBI Exome Sequence Project. JAMA Neurol. 72, 781–788 (2015).
Cheng, S. et al. Exome-wide screening identifies novel rare risk variants for major depression disorder. Mol. Psychiatry 27, 3069–3074 (2022).
Palmer, D. S. et al. Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat. Genet. 54, 541–547 (2022).
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Jurgens, S. J. et al. Analysis of rare genetic variation underlying cardiometabolic diseases and traits among 200,000 individuals in the UK Biobank. Nat. Genet. 54, 240–250 (2022).
Zhou, W. et al. SAIGE-GENE+ improves the efficiency and accuracy of set-based rare variant association tests. Nat. Genet. 54, 1466–1469 (2022).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Fong, T. G. & Inouye, S. K. The inter-relationship between delirium and dementia: the importance of delirium prevention. Nat. Rev. Neurol. 18, 579–596 (2022).
Davies, G. et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 9, 2098 (2018).
Weiner, D. J. et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614, 492–499 (2023).
Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 9, 5269 (2018).
Patsopoulos, N. A. et al. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Meier, S. M. et al. Genetic variants associated with anxiety and stress-related disorders: a genome-wide association study and mouse-model study. JAMA Psychiatry 76, 924–932 (2019).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Arnold, P. D. et al. Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Ruderfer, D. M. et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715.e1716 (2018).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Garcia, F. J. et al. Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e214 (2019).
Català-Senent, J. F. et al. A deep transcriptome meta-analysis reveals sex differences in multiple sclerosis. Neurobiol. Dis. 181, 106113 (2023).
Wendler, F., Purice, T. M., Simon, T., Stebbing, J. & Giamas, G. The LMTK-family of kinases: emerging important players in cell physiology and pathogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 165372 (2021).
Cheng, X. R., Zhou, W. X. & Zhang, Y. X. The behavioral, pathological and therapeutic features of the senescence-accelerated mouse prone 8 strain as an Alzheimer’s disease animal model. Ageing Res. Rev. 13, 13–37 (2014).
Shribman, S., Reid, E., Crosby, A. H., Houlden, H. & Warner, T. T. Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches. Lancet Neurol. 18, 1136–1146 (2019).
Lim, J. H. et al. Missense mutation of SPAST protein (I344K) results in loss of ATPase activity and prolonged the half-life, implicated in autosomal dominant hereditary spastic paraplegia. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3221–3233 (2018).
Mészárosová, A. U. et al. SPAST mutation spectrum and familial occurrence among Czech patients with pure hereditary spastic paraplegia. J. Hum. Genet. 61, 845–850 (2016).
Verriello, L. et al. Amplifying the spectrum of SPAST gene mutations. Acta Biomed. 92, e2021220 (2021).
Serrano-Pozo, A., Das, S. & Hyman, B. T. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 20, 68–80 (2021).
Bramall, A. N., Anton, E. S., Kahle, K. T. & Fecci, P. E. Navigating the ventricles: novel insights into the pathogenesis of hydrocephalus. EBioMedicine 78, 103931 (2022).
Mercuri, E., Pera, M. C., Scoto, M., Finkel, R. & Muntoni, F. Spinal muscular atrophy—insights and challenges in the treatment era. Nat. Rev. Neurol. 16, 706–715 (2020).
Lioutas, V. A. et al. Incidence of transient ischemic attack and association with long-term risk of stroke. JAMA 325, 373–381 (2021).
Romero, C. et al. Exploring the genetic overlap between twelve psychiatric disorders. Nat. Genet. 54, 1795–1802 (2022).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019).
Grotzinger, A. D. et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat. Genet. 54, 548–559 (2022).
Szustakowski, J. D. et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 53, 942–948 (2021).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Vaser, R., Adusumalli, S., Leng, S. N., Sikic, M. & Ng, P. C. SIFT missense predictions for genomes. Nat. Protoc. 11, 1–9 (2016).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. https://doi.org/10.1002/0471142905.hg0720s76 (2013).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Nagel, M., Watanabe, K., Stringer, S., Posthuma, D. & van der Sluis, S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 9, 905 (2018).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Watanabe, K., Umićević Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019).
Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31, 968–980 (2006).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Inker, L. A. et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N. Engl. J. Med. 367, 20–29 (2012).
Wu, B. Summary data for ‘Single variant analysis for common variants’. Figshare https://doi.org/10.6084/m9.figshare.25342486.v1 (2024).
Wu, B. Summary data for ‘Exome-wide gene-based association analyses’. Figshare https://doi.org/10.6084/m9.figshare.25342201.v1 (2024).
Acknowledgements
We thank all the participants and researchers from the UKB. We also thank the SynGO Consortium. This study was supported by grants from the STI2030-Major Projects (no. 2022ZD0211600 to J.-T.Y.), the National Natural Science Foundation of China (nos 82071201, 82271471 and 92249305 to J.-T.Y.; no. 82071997 to W.C.), the Shanghai Municipal Science and Technology Major Project (no. 2018SHZDZX01 to J.-F.F.), the Research Start-up Fund of Huashan Hospital (no. 2022QD002 to J.-T.Y.), the Excellence 2025 Talent Cultivation Program at Fudan University (no. 3030277001 to J.-T.Y.), Shanghai Talent Development Funding for the Project (no. 2019074 to J.-T.Y.), the Shanghai Rising-Star Program (no. 21QA1408700 to W.C.), the 111 Project (no. B18015 to J.-F.F.), the National Key Research and Development Program of China (no. 2023YFC3605400 to W.C.), the Postdoctoral Innovation Talents Support Program (no. BX20230087 to S.-D.C.; no. BX20230089 to Y.-R.Z.) and ZHANGJIANG LAB, Tianqiao and Chrissy Chen Institute, the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, and the Shanghai Center for Brain Science and Brain-Inspired Technology, Fudan University. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors had full access to the data in the study and accepted responsibility to submit it for publication. J.-T.Y. designed the study. Y.-T.D. and B.-S.W. conducted the primary analyses and drafted the manuscript. L.Y., J.-J.K., W.-S.L., Z.-Y.L., X.-R.W. and X.-Y.H. contributed to the biological annotation analyses and brain structures, proteomics and inflammatory marker association analyses. J.-T.Y., W.C., Y.Z., J.-F.F., Y.M., Q.D., Y.-R.Z., S.-D.C., Y.-J.G. and Y.-Y.H. critically revised the manuscript, and all authors approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Human Behaviour thanks Laura Fahey, Hei Man Wu and and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Burden heritability of neuropsychiatric diseases and traits.
a. The burden heritability of neuropsychiatric diseases calculated by burden heritability regression (Methods). The x-axis indicates the specific neuropsychiatric disease and the y-axis indicates the heritability based on rare variants. b. The burden heritability of neuropsychiatric traits calculated by burden heritability regression (Methods). The x-axis indicates the specific neuropsychiatric disease and the y-axis indicates the heritability based on rare variants. c. The distribution of heritability based on different annotation groups in neuropsychiatric diseases (n = 46). Each color represents one annotation group as indicated in the plot legend. Horizontal bars indicate median value and rhombuses indicate mean value of the burden heritability. The lower and upper hinges represent the 25th and 75th percentiles. The upper whisker extends from the hinge to the largest value no further than 1.5 times the inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5*IQR of the hinge. The data beyond the end of the whisker is plotted separately, in which case the minimum and maximum values are the bottom and top points respectively, while if there are no separate points drawn, the minimum and maximum values are the end of the lower and upper whisker respectively. The sample sizes for each disease were provided in Supplementary Table 1,c. d. The distribution of heritability based on different annotation groups in neuropsychiatric traits (n = 23). Each color represents one annotation group as indicated in the plot legend. Horizontal bars, hinges and whiskers indicate same type of statistics as c. The sample sizes for each disease were provided in Supplementary Table 1,d.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Tables 1–17.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, YT., Wu, BS., Yang, L. et al. Large-scale whole-exome sequencing of neuropsychiatric diseases and traits in 350,770 adults. Nat Hum Behav (2024). https://doi.org/10.1038/s41562-024-01861-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41562-024-01861-4