Abstract
Allorecognition, the capacity to discriminate self from conspecific non-self, is a ubiquitous organismal feature typically governed by genes evolving under balancing selection. Here, we show that in the fungus Podospora anserina, allorecognition loci controlling vegetative incompatibility (het genes), define two reproductively isolated groups through pleiotropic effects on sexual compatibility. These two groups emerge from the antagonistic interactions of the unlinked loci het-r (encoding a NOD-like receptor) and het-v (encoding a methyltransferase and an MLKL/HeLo domain protein). Using a combination of genetic and ecological data, supported by simulations, we provide a concrete and molecularly defined example whereby the origin and coexistence of reproductively isolated groups in sympatry is driven by pleiotropic genes under balancing selection.
Similar content being viewed by others
Main
The capacity to discriminate self from non-self occurs throughout the Tree of Life1,2,3 and is necessary for fundamental processes such as multicellular growth, detection of pathogens and choice of mating partners. Genetically, the detection of non-self is achieved by the product of highly polymorphic genes that are subject to various forms of balancing selection, in particular negative frequency dependence where rare alleles have a fitness advantage4,5,6,7,8. Since balancing selection ensures the coexistence of several variants in a population, it can be hypothesized that self/non-self recognition genes with pleiotropic interactions on sexual reproduction could lead to the evolution of reproductively isolated groups and hence speciation. For instance, the major histocompatibility complex (MHC), which is an essential element of the adaptive immune system in jawed vertebrates, appears to contribute to and accelerate speciation by favouring assortative mating9,10,11. Likewise, in organisms such as fungi, slime moulds and colonial marine invertebrates, vegetative incompatibility (allorecognition) systems might lead to reproductive isolation if they display pleiotropic effects on sexual reproduction.
Here, we address this hypothesis directly using the model fungus Podospora anserina, for which the allorecognition system is well-studied12,13,14. As in other fungi, vegetative fusion between individuals of P. anserina is controlled by the so-called het (for heterokaryon incompatibility) genes. Fusion within a fungal mycelium is generally regarded as advantageous for cytoplasmic transport and colony establishment15,16. However, fusion between different individuals can be deleterious since it allows for the transmission of viruses, defective plasmids and selfish nuclei17,18,19,20,21. It is believed that the primary function of the het genes is to avoid such risks, as successful fusion is only possible if individuals are compatible at all of their het genes, otherwise triggering regulated cell death of the fused cell22. It has also been proposed that some het genes are involved in pathogen recognition and that the vegetative incompatibility is a secondary by-product of their evolution23,24. In that sense, the het genes can be seen as analogous to components of the innate immune system of animals and plants24. Notably, the genetic basis of compatibility is generally different between the vegetative and sexual stage but some het genes are known to have pleiotropic effects on the sexual cycle of a number of species25,26. Of the nine genetically identified het loci in P. anserina, six are known to have antagonistic pleiotropic effects on the sexual function through sterility or progeny inviability12,27 (Supplementary Fig. 1). In this study, we used population genomic analyses, laboratory crosses, genetic manipulations, field observations and simulations to show that two het genes define reproductively isolated groups in P. anserina and propose scenarios of how this arises.
Results
Low genetic diversity and balancing selection in P. anserina
We started by sequencing the haploid genomes of 106 P. anserina strains, spanning 25 years of sampling around Wageningen, the Netherlands (from 1991 to 2016; Supplementary Table 1). Whole genome sequence data showed that the P. anserina samples are remarkably similar, with an average pairwise nucleotide diversity (π) of 0.000492 (Fig. 1a and Supplementary Fig. 2). As P. anserina does not produce asexual propagules, we expect clonality to be low but the extremely low genetic diversity could also be the result of very high selfing rates and demographic processes like bottlenecks. As an indication of the degree of outcrossing, we calculated the linkage disequilibrium (LD) decay for the Wageningen collection (Supplementary Fig. 3). We found that LD, as measured by the r2 statistic, reaches 0.2 at distances <3.5 kilobases (kb) for most chromosomes, except for chromosome 4 that has a much slower decay (r2 < 0.2 after 12.2 kb; Methods). These values are intermediate between those typical for outcrossers and for extreme selfers in fungi28. Additionally, we sampled 68 new genetically different strains in 2017 around Wageningen (Supplementary Table 2; Methods) and investigated the occurrence of spore killing, a phenotypic expression of meiotic drive in fungi. Previous studies have shown that natural populations of P. anserina harbour a number of meiotic drivers (selfish genetic elements that cause segregation distortion) belonging to the Spok gene family29,30. Mating between an individual with a meiotic driver and an individual without it results in the abortion of the spores that did not inherit the meiotic driver. Hence, observing spore killing is a direct indication of an outcrossing event29. We encountered one case of spore killing in our sample despite the low frequency of active spore killers in this population (<20%; Supplementary Table 1)29. Taken together, we conclude that, while showing indications of a high selfing rate, P. anserina outcrosses at detectable levels in nature.
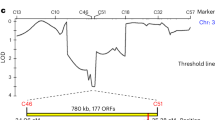
a, Sliding window analysis (10 kb long with steps of 1 kb) of representative chromosomes 1 (top) and 3 (bottom) with values of genetic diversity (measured as either the pairwise nucleotide diversity π in red or as Watterson’s theta θW in blue) and the Tajima’s D statistic (orange). The location of relevant loci is marked when present: het genes, the centromere, the mating type locus (MAT) and the location of meiotic drivers of the Spok family. Loci with highly divergent alleles (for example, MAT) and repetitive regions (for example, WD40 repeats of HNWD genes) were filtered out by the variant-calling pipeline but linked variants can still show signals of balancing selection. Note that Spok3 and/or Spok4 can be found at different locations in the genome depending on the strain. b, Changes of allele frequencies through time for three known het loci (data shown from years with more than five samples).
In contrast with the observed general low genetic diversity, previous studies have found a great variety of vegetative incompatibility groups (defined by the het genes) within the P. anserina Wageningen collection31,32. Under the expectation that the het genes evolve under negative frequency-dependent selection33,34,35, we calculated the Tajima’s D statistic in sliding windows along the genome (Fig. 1a and Supplementary Fig. 2). As expected, the windows containing most het genes exhibit high genetic diversity and locate directly under high positive Tajima’s D peaks (Fig. 1a and Supplementary Fig. 2), an indication of balancing selection. The het genes that do not colocalize with positive Tajima’s D values belong to the HNWD gene family (for example, het-d and het-r in chromosome 2; Supplementary Fig. 2), which are characterized by WD40 tandem repeats at the C terminus that define the allele specificity36,37. The repetitive end of the HNWD genes excluded them from our variant-calling pipeline, making genome scans inappropriate to assess signatures of selection for such genes. Nonetheless, other members of the HNWD family (for example, het-e and hnwd3 in chromosomes 4 and 7, respectively; Supplementary Fig. 2) are flanked by sites with high Tajima’s D, suggesting that at least some members of this gene family evolve under balancing selection.
To complement the signatures of historical demographic processes provided by the Tajima’s D statistic, we took advantage of our temporal data and plotted the changes in allele frequencies through time for representative het genes in the Wageningen samples (Fig. 1b and Supplementary Fig. 4). As expected under balancing selection, the het genes show intermediate allele frequencies over the sampling time period. This pattern includes the het-c gene (Fig. 1b), which is the interacting partner of het-d and het-e38, providing further support for the hypothesis that HNWD genes are also under balancing selection31.
Two reproductively isolated groups exist within P. anserina
Previously, van der Gaag39 performed pairwise crossings of around half the Wageningen collection (plus the French reference strain S), recording various degrees of sexual and vegetative incompatibility. To gain insight into the population structure, we re-coded these observations into a distance matrix of mating success for 45 strains, for which we also have Illumina data (Supplementary Table 3). A principal coordinates analysis (PCoA) of the distance matrix revealed two groups, as identified by a partitioning around medioids (PAM) clustering method (Fig. 2a). Notably, the same two groups can be identified using a principal component analysis (PCA) of whole-genome single nucleotide polymorphism (SNP) data (Supplementary Fig. 5a). Moreover, analysis by chromosome reveals that the clustering signal is driven by variation on chromosome 5 (Fig. 2a) and to a lesser extent chromosome 2 (Supplementary Fig. 5b–d).
a, Left, a PCoA of mating success data reveals two groups (indicated by lilac and green ovals); right, a PCA of the SNP data from chromosome 5 returns clusters of the same two groups (divided by the grey line). Samples with both mating and genomic data are coloured on the basis of the mating success clustering (lilac and green points). Samples without mating success data are in grey. b, Genetic differentiation between the two groups (as defined by the PC1 axis of the SNP data) shown as Fst and DXY statistics estimated from sliding windows (10 kb long with steps of 1 kb).
On the basis of the result of the clustering analysis, we divided all the samples into what we refer to hereafter as the two ‘reproductively isolated’ (RI) groups. For samples with no mating success data (59.8% of the total), we assumed RI group membership on the basis of the first principal component of chromosome 5 (Fig. 2a). We computed the population fixation index Fst and the divergence statistic Dxy between the RI groups in windows along each chromosome (Fig. 2b, Supplementary Fig. 6 and Supplementary Fig. 7). While most chromosomes show virtually no differentiation, we found that a region near the centre of the left arm of chromosome 5 is strongly differentiated (nearly reaching the maximum Fst = 1 and having the highest Dxy value). In accordance with the PCA, a region on chromosome 2 also shows intermediate to high levels of differentiation (on the basis of Fst but not of Dxy). The area of differentiation in chromosome 2 co-localizes with the het-r gene, one of the het genes with pleiotropic effects on sexual reproduction (Supplementary Fig. 1). From classical genetic analyses, it is known that het-r works as an allorecognition gene through its interaction with the uncharacterized het-v locus12, which is localized on chromosome 5 (ref. 40). Hence, although other het genes cause sexual incompatibilities, this interacting pair is the most likely functional candidate behind the partition into two RI groups and the associated genomic divergence. We thus focused specifically on these two loci in further analyses.
Like other members of the HNWD family, the het-r gene has a WD40 repeat domain at its C terminus37. The number of tandem WD40 repeats and their sequence have been shown to define two alleles: R and r (ref. 36). Allele R has 11 WD40 repeats in a specific order. In effect, allele r is a non-functional variant that can have any other number of repeats or a mutated version of the basic 11 repeats36. The het-v locus also has two alleles, V and V1, as defined by classical genetics41. The vegetative incompatibility reaction (or barrage) is triggered when individuals of different het-v alleles meet and fuse (Fig. 3a). Different het-r alleles do not trigger barrage formation on their own but there is a non-allelic interaction between the alleles R and V leading to a barrage. Crucially, the allelic and non-allelic vegetative interactions are mirrored by sexual sterility with severity depending on the genotype of the parents42,43. Moreover, individuals of the genotype RV are self-incompatible, making that combination lethal upon germination41,44 (Fig. 3a). While the het-r/v incompatibility had been previously linked to sexual dysfunction, there had been no indication of this leading to two RI groups as observed here. To confirm that het-r/v are indeed responsible for causing RI in P. anserina, we molecularly characterized the het-v locus.
a, When two individuals with incompatible alleles fuse during vegetative growth, an incompatibility reaction occurs producing a divisive line of dead cells known as barrage. During sex, when the female organ (protoperithecium) is fertilized by microconidia, different genotypes of the two het genes confer various degrees of infertility depending on the crossing partner. The RV genotype is lethal. b, The location of het-v was identified through nested deletions, marked in orange if they lost the barrage formation in confrontations with a V1 strain (that is, became compatible) or in grey if they did not. The black bar represents a cluster of genes absent in the strain Y, which is of incompatibility type V1. c, Deletion Δ113 (Δ12915-123170) from a V strain leads to the V1 phenotype. A barrage test with a V (top row) and V1 tester (bottom row) is given for three transformants obtained with the Δ113 deletion cassette using an rV recipient strain. The central transformant t2 produces a barrage reaction to V but not to V1 (that is, it acquired the V1 phenotype). Transformants t1 and t3 retain the V phenotype and are presumably V + V1 heterokaryons. d, Genetic structure of the het-r and het-v loci is represented by the domain architecture of the encoded proteins. Arrows represent incompatible interactions. e, To the left, a cross between rV (light colour) and RV1 (dark colour) produces very few fruiting bodies. However, fertility is recovered by deleting the het-v locus and adjacent genes (Δ12810-12690) as shown in the cross in the middle. Reintroduction of the het-Va and het-Vb genes fully restores the sterile phenotype characteristic of rV × RV1 in the cross to the right.
Locus het-v encodes two genes that cause incompatibilities
As the het-v locus was genetically mapped to the left arm of chromosome 5, we introduced genetic markers in that region and analysed their linkage to the vegetative incompatibility phenotype of het-v (Supplementary Fig. 8; Methods). This led to the identification of a 113 kb long candidate region (Fig. 3b). Deletion of this region in an rV strain led to loss of the barrage reaction to an otherwise isogenic rV1 strain, as expected if het-v locates there. Notably, while losing the V phenotype, these deletion strains simultaneously acquired the V1 phenotype (rather than showing a neutral incompatibility phenotype) (Fig. 3c). In other words, deleting the region with the V allele resulted in vegetative incompatibility to V and compatibility to V1.
Using additional nested deletions in the rV strain (Fig. 3b and Supplementary Fig. 8) we narrowed down the candidate region to a 38 kb area (between the genes Pa_5_12810 and Pa_5_12690, henceforth Δ12810-12690; ref. 45 gives gene codes). Inspection of the sequenced genome of the wild-type strain Y, which is of the V1 incompatibility type, revealed that a ~12 kb region spanning five genes (Pa_5_12750 to Pa_5_12710) is replaced by a cluster of transposable elements when compared with the reference genome of the S strain45, which is of the V type. Consistent with the deletion strain experiment, these five genes are contained within the Δ12810-12690 area. The V1 phenotype of the deletion strains, in turn, indicates that the cluster of transposable elements at the wild-type V1 locus is not required for the V/V1 incompatibility.
To determine which of the five open reading frames (ORFs) within the region absent in the strain Y are responsible for the V phenotype, we turned to a complementation cloning approach using the rV Δ12810-12690 strain (which displays the V1 phenotype) as a recipient. Our results showed that insertion of the genes Pa_5_12710 and Pa_5_12720 confers the V phenotype on the recipient strain and that these two genes thus correspond to het-v. Furthermore, both genes together are required for allelic incompatibility to V1, while Pa_5_12720 alone determines non-allelic incompatibility to R (Fig. 3d and Supplementary Table 4). A strain expressing Pa_5_12720 (but not Pa_5_12710) becomes compatible with both V and V1. Thus, we propose to name Pa_5_12720 and Pa_5_12710 as het-Va and het-Vb, respectively (Fig. 3d).
Since het-Va encodes a predicted lysine or histidine methyltransferase (Fig. 3d and Supplementary Fig. 9), we hypothesized that this activity was required for the incompatibility function. Two different point mutants of the catalytic tyrosine residue were obtained (Y233A and Y233F; Supplementary Fig. 9). When introduced into a recipient with the rV1 phenotype (V Δ12810-12690), both mutants restored vegetative incompatibility to R (Supplementary Table 4). In contrast, co-transformation of het-Vb with het-Va Y233A or Y233F failed to restore the barrage reaction to V1. We conclude that the methyltransferase activity of HET-Va is required for the allelic V1/V vegetative incompatibility but dispensable for non-allelic R/V vegetative incompatibility (Fig. 3d).
To confirm that the vegetative and sexual effects associated with het-v have the same mechanistic basis, we verified that the Δ12810-12690 deletion of the region encompassing het-Va and het-Vb, which converts V to the V1 vegetative phenotype, also restores fertility in crosses to an RV1 strain (Fig. 3e and Supplementary Fig. 10). We found that a Δ12810-12690 × RV1 cross shows normal fertility. Moreover, when het-Va and het-Vb are re-introduced by transformation into Δ12810-12690 strains, the crosses to an RV1 strain show sterility. In addition, we found concordance between the sexual and vegetative phenotypes when the het-Va point mutants were used for transformation (Supplementary Fig. 11 and Supplementary Table 5). An rV1 recipient transformed with het-Va Y233F (or Y233A) together with het-Vb showed normal (female) fertility to rV1, while transformants expressing het-Va Y233F remained sterile (as males) in crosses to RV1. Thus, as for the vegetative incompatibility, the methyltransferase activity is dispensable for R/V but required for V/V1 sexual incompatibility. However, a discrepancy between the vegetative and sexual effects was noted for the het-Va Y233A mutation, which suppressed R/V sexual but not vegetative incompatibility. It is possible that this less conservative substitution partially destabilizes the het-Va product and that decreased protein amounts are sufficient to trigger vegetative but not sexual incompatibility. Accordingly, it has been observed in other P. anserina het systems that higher protein amounts are required for sexual than for vegetative incompatibility46. Thus, we conclude from these experiments that het-Va and het-Vb are responsible both for the vegetative and sexual phenotype of het-v and that sexual incompatibility is not the result of linked variants.
In essence, because the cell death reaction is not turned off during sexual reproduction, the het-r/v system acts as a reproductive barrier (Fig. 4; also Supplementary Fig. 12). Asymmetry in fertilization probably arises from the fact that the V allele is a diffusible cytoplasmic factor, while the HET-R protein is not41,43. Potentially, the HET-R protein is found in very low amounts or does not diffuse from the male gamete (microconidium) to the female organ (protoperithecium). As a result, the R protoperithecium is damaged by the V products but not vice versa (Fig. 4c).
a, Vegetative cell fusion between members of the two RI groups is prevented by the allelic interaction of the het-v alleles (V/V1) and the non-allelic interaction between het-v and het-r (V/R), which deters the transfer of viruses and other deleterious cytoplasmatic elements. b, During sex, the het-r/v system remains active, resulting in failed fertilization. c, In most cases the zygote formation does not occur and female organs (protoperithecia) do not develop into perithecia. The few successful perithecia produce spores but a fraction of these spores are self-incompatible and die 15 h after germination12,41,42,43,44. While female R organs are irreversibly damaged after abortive fertilization with V microconidia, female V organs are still viable after fertilization with R (or V1) microconidia41,43.
In combination with previous knowledge on het-r, we can now derive a mechanistic model for het-r/v allorecognition function. It is known that HET-R has a tripartite domain organization typical of nucleotide-binding oligomerization domain (NOD)-like receptors or NLRs23,37. Thus, the het-r/v interaction can be hypothesized to function analogously to other incompatibility systems involving NLRs such as het-c/e/d, het-z and the het-s/nwd2 systems31,47,48. In such a model, HET-R would bind the HET-Va protein via the variable C-terminal domain of WD40 repeats, which would trigger oligomerization of its NACHT domain and downstream activation of the HET cell death-inducing domain. This model accounts for the fact that WD40 repeat loss in HET-R leads to the inactive r phenotype36 and that R/V incompatibility is independent of the predicted methyltransferase activity of HET-Va.
The genetics of the V/V1 incompatibility, by contrast, represents a rather puzzling situation from a mechanistic point of view. It is unclear how the het-Va and het-Vb genes (that is, the V allele) are able to sense the absence of their own products (V1) during the vegetative (and sexual) incompatibility reaction. HET-Va is a predicted methyltransferase, while HET-Vb displays a TUDOR domain occurring in so-called reader proteins, which recognize the methylation marks deposited by methyltransferases49, as well as a MLKL/HeLo membrane-targeting cell death-inducing domain50. The domains involved suggest that incompatibility could be brought about by a combination of methyl mark deposition (by HET-Va) and reading by the HET-Vb TUDOR domain and subsequent activation of the MLKL/HeLo domain ensuring cell death execution (Supplementary Fig. 13), by analogy to the mechanism of cell death execution in mammalian necroptosis or het-s incompatibility50.
At any rate, regardless of the mechanism of V/V1 incompatibility, the implication of the HET-R NLR stands out because NLRs are key components of the innate immune system of plants and animals23,51,52. Immunity genes, like the het genes, are expected to evolve under negative frequency-dependent selection, although driven by pathogen pressure instead of vegetative fusion. Bateson–Dobzhansky–Muller incompatibilities53 can arise between populations adapted to different pathogen pools, which in plants can result in the autoimmune response known as hybrid necrosis54. Crucially, characterized hybrid necrosis genes happen to be NLRs themselves in, for example, Arabidopsis thaliana55, tomato56 and cotton57. Thus, genes of similar molecular characteristics and selective pressures driven by non-self recognition can contribute to reproductive isolation in vastly different taxa.
The het-r and het-v loci induce reproductive isolation in the wild
Once the molecular basis of vegetative and sexual incompatibility was identified, we set out to further characterize the Wageningen population with regard to the two RI groups. Although the effects of the het-r/v interaction on sexual incompatibility are significant, they are not absolute. Hybrids can be produced in the laboratory (Fig. 3e, left) and the RV reaction is thermosensitive, losing effect at 32 °C (ref. 58). To evaluate if the RI group effects we observed in the laboratory crosses hold in wild populations, we analysed the population genomic dataset to determine their het-r/v alleles. We reason that if, despite sexual incompatibilities, the two RI groups are mixing in nature, we should encounter the three possible viable genotypes between het-v and het-r (that is, rV, RV1 and the recombinant rV1; Fig. 3a) in the wild, simply because of independent chromosomal segregation. We confirmed that the RI groups (as in PC1 in Fig. 2a) are perfectly defined by the het-v allelic identity. Indeed, the maximum values of Fst occur at the het-v locus (Fig. 2b and Supplementary Fig. 6). Since the members of the HNWD family cannot be assembled with Illumina data, we used a PCR-based method of ref. 59 to genotype the het-r gene in the Wageningen collection (Supplementary Tables 1 and 6). As expected, under strong reproductive isolation, we found a nearly perfect association of r with V and R with V1 throughout the sampled years (Fig. 5a), with an LD estimate of r2 = 0.926 between the two loci. By contrast, het-v has no association with other het genes, such as het-z in chromosome 1 (r2 = 0.004) or het-q in chromosome 7 (r2 = 0.009), showing that the LD is not genome-wide (Supplementary Fig. 14a). We further found a modest association of sites around (linked to) het-v and the area of chromosome 2 surrounding het-r (Supplementary Fig. 14b), further supporting the association between the het-r/v genotypes.
a, Genotypes of het-v and het-r in the Wageningen population through time. The rV1 genotype is very rare, confirming that the RI groups (RV1 and rV) are not mixing freely. RV is lethal. b, There is no significant difference (NS) in abundance between RI groups based on the source herbivore (Pearson’s Χ21, 82 = 0.46365, P = 0.4959; genotyped samples from 1991 to 2016 with substrate data, n = 82). Sheep dung was not tested due to low sample sizes. c, The strains collected in 2017 from horse dung (plus one sample from sheep) show that the RI groups can be found in the same dung piece (n = 63). Insert, fruiting body of P. anserina in the wild (photo by S.L.A.V.).
One may hypothesize that the lack of mixing between RI groups is strengthened by differences in ecology, such as substrate or temporal isolation. Like other coprophilous fungi, P. anserina has a short life cycle (completed in around 11 d under laboratory conditions), where the sexual spores are ejected into the surrounding vegetation and subsequently ingested by herbivores. The spores go through the animal’s digestive tract and germinate, grow and reproduce within the dung (Supplementary Fig. 15). From the genotyped strains with substrate information available, we saw no significant differences in the abundance of each RI group (for horse and rabbit dung: Pearson’s Χ21, 82 = 0.46365, P = 0.4959; Fig. 5b). To assess the possibility of temporal isolation, we phenotypically assigned strains collected in 2017 to each RI group by confronting them with tester strains of known genotype. Remarkably, the RI phenotyping matched perfectly the expected het-v allele in a subset of samples evaluated with PCR (Supplementary Table 2). We simultaneously confronted strains sampled within the same dung piece to each other and used barrage formation to estimate the number of distinct individuals present. We found that more than one individual can be found in the same dung piece and that such individuals often belong to different RI groups (Fig. 5c). Taken together, this demonstrates that the RI groups do not lack opportunities for mixing in the wild.
Simulations recapitulate the formation of two RI groups
To investigate the conditions under which a split of a population by means of pleiotropic genes under balancing selection can occur, we designed an individual-based simulation using SLiM60 on the basis of the Podospora life cycle and our characterization of the het-r/v interaction (Supplementary Methods and Supplementary Fig. 16). Since we do not know the ancestral state of either het gene, we used as starting point populations of a single genotype, either rV or RV1, and then a small fraction of individuals of opposite genotype were introduced. Regardless of the starting genetic composition, we found that balancing selection and some degree of selfing is required for the introduced genotype to invade the population (Fig. 6 and Supplementary Figs. 17 and 18). Selfing is needed because the invader allele immediately suffers the cost imposed by the lethal RV combination produced during outcrossing, which will prevent it from increasing in frequency. In the presence of selfing, the cost can be avoided and balancing selection then provides benefits to the invader alleles through rare-allele advantage, eventually driving them to intermediate frequencies. Importantly, the population is composed of the two expected genotypes (rV and RV1, to the exclusion of rV1) under several scenarios, most of them dependent on the strength of balancing selection acting on the non-allelic (R/V) system and secondarily on the allelic one (V/V1) (Fig. 6 and Supplementary Figs. 17 and 18). In addition, high levels of selfing, prezygotic isolation or both, can lower the frequencies of the rV1 genotype, completing isolation. Notably, a theoretical study on hybrid necrosis genes in plants also found a role of negative frequency dependence on the build-up of reproductive barriers61, with the difference that the isolating barrier in question is exclusively postzygotic, instead of prezygotic and postzygotic as in the case of P. anserina. Their model also included selfing but the authors did not explore the effect of this parameter on their results. Overall, we conclude from the simulations that RI groups can evolve from pleiotropic allorecognition loci in organisms that undergo selfing (or inbreeding) at high rates during the invasion of one of the two incompatible genotypes. Analogous to the scenario where pleiotropic effects of the MHC drive speciation through assortative mating11, in Podospora selfing acts as the non-random mating process that allows reproductive isolation. Moreover, sympatry is a necessary condition for the evolution of reproductive isolation in our model, since vegetative interactions between individuals of the two groups are required for balancing selection to operate.
For each parameter combination, the distribution of genotype frequencies of 100 replicated simulations is given. Each simulation is represented by a dot and their distribution by a boxplot, which shows the median as a diamond, the 25th to 75th percentiles as the box bounds and 1.5× interquartile ranges as whiskers. If dots and boxplots are not visible, the distribution is concentrated behind the diamond of the median.
Of note is the fact that, out of all the het systems of P. anserina that have effects on the sexual function, only het-r/v leads to detectable effects on population structure and compatibility type differentiation. Both the het-c/e/d and the het-z systems lead to sterility in specific combinations of het-genotypes in the male and female parent (Supplementary Fig. 1)12. Likewise, the unlinked loci het-c/e/d can produce ‘self-incompatible’ progeny that die after germination, just like het-r/v12,42. However, our simulations show that, in the absence of balancing selection acting on an allelic system and the associated prezygotic incompatibilities (roughly equivalent to a simplified het-c/d or het-c/e interaction, for example), the two mating groups can only evolve under extremely high selfing rates. Probably, the fact that sterility is (nearly) symmetrical in the het-r/v system, but not in the other ones, makes it particularly prone to split the population.
Admittedly, our simulations do not fully capture the complexity of the multiallelic het-c/e/d system, where sexual incompatibility can be additive. For instance, a C1E1 × C2E2 cross is nearly totally sterile, as their het-c and het-e alleles are incompatible. Yet, there are numerous wild-isolates that show neutral het-d and het-e alleles12. Presumably, the strains with neutral het-d and het-e alleles can bridge the gene flow between incompatible isolates, limiting their effects on reproductive isolation. Interestingly, the het-c alleles that can interact with more het-e/d alleles and which would be more effective at inducing reproductive isolation, have a low frequency in the Wageningen population (alleles C4, C8 and C9 in Fig. 1), suggesting that they are too costly relative to their adaptive value31.
The het-v locus is physically close to a meiotic drive gene
In addition to the factors discussed above, meiotic drive could further influence the maintenance of allorecognition genes with pleiotropic effects. The advantage provided by segregation distortion can counteract selection against deleterious alleles, leading to stable polymorphism62. We genotyped all samples for the presence of members of the Spok family (Supplementary Table 1), confirming previous indications29 that the Spok2 gene is at a high population frequency (86.7%). The location of Spok2 is relatively close to het-v and falls within the area of significant differentiation between RI groups (Supplementary Figs. 6 and 14). If the presence of Spok2 predates the evolution of het-r/v, the segregation distortion advantage of Spok2 could have aided the establishment of the incompatibility system despite the cost of sexual incompatibility. Alternatively, Spok2 could have invaded P. anserina after the evolution of het-r/v. As spore killers generally only experience the fitness boost from meiotic drive after reaching some minimum population frequency63, Spok2 may have benefited from hitchhiking along with a locus under balancing selection at early stages of population invasion. Recombination could have later decoupled Spok2 from a given het-v allele, allowing it to invade both RI groups and approach fixation. The invasion of Spok2 could have then eroded divergence between RI groups, as observed in meiotic drive systems of Drosophila species64. Data from different populations of Podospora are required to clarify this link but, regardless, the proximity of Spok2 and het-v hint to a previously unappreciated way meiotic drive could contribute to speciation.
The het-r and het-v loci are present in related species
Finally, in an attempt to discern the evolutionary history of het-r and het-v in P. anserina, we examined the genomes of other members from the P. anserina species complex65. We found that orthologues of het-r are present in all seven species of the complex, although their allelic identity is unknown (Methods). In the case of het-v, we found that het-Va and het-Vb are present in at least some species but there is also evidence of introgression of het-v between P. anserina and P. pauciseta (Supplementary Fig. 19). These observations suggest a dynamic evolution of the het-v locus throughout the divergence of this species complex but without population sampling from the other Podospora species we cannot draw concrete conclusions.
Looking at other genera of the order Sordariales66, we also found that het-Va and het-Vb show conserved synteny in Sordaria macrospora and a number of Neurospora species (Supplementary Table 6). As large genomic datasets of the Neurospora species are available, we investigated the distribution of the het-v alleles across the genus. We found that only Neurospora perkinsii, N. intermedia and some lineages of N. tetrasperma have full copies of both het-Va and het-Vb. In N. crassa and N. sitophila, the gene het-Vb seems pseudogenized, while in N. metzenbergii and N. tetrasperma lineages L4, L7 and L8 (ref. 67) both genes are completely absent. Notably, N. intermedia and some lineages of N. tetrasperma are polymorphic for pseudogenized and full copies of het-Vb, raising the question of whether het-v also works as an allorecognition gene in these species. Furthermore, different taxa have their own premature stop codons, suggesting independent loss events. Thus, although we lack mechanistic evidence for the function of these genes in Neurospora, the observed patterns suggest that het-v might be old but involved in rapid presence/absence turnover in other taxa. By contrast, we found that het-r gene does not appear to be present in Neurospora.
Importantly, the het-r/v interaction itself cannot be ancestral to the P. anserina species complex, as the het-r/v incompatibility leads to speciation and thus to the loss of polymorphism (that is, each genotype is ‘fixed’ in each RI group). Accordingly, the extremely low diversity and the lack of differentiation along the genome (based on both Fst and Dxy), other than around het-v and het-r, suggest that the RI group isolation is extremely recent. In addition, as it is known that there is a thermosensitive component to the R/V interaction44, it remains of interest to study the phylogeography of this incompatibility system.
Conclusion
Dissecting the genetic basis of reproductive barriers is a major challenge in speciation research68,69. In P. anserina we found that the het-r/v system not only contributes to speciation but it directly defines RI groups by conflating vegetative recognition with sexual compatibility. If current conditions are maintained, the P. anserina RI groups may accumulate further genetic incompatibilities and evolve ecological differences, eventually completing the speciation process. While the typical number and effect sizes of speciation genes (sensu ref. 70) at the onset of speciation remains an open question, the case of this fungus suggests that speciation can happen through few loci of big effects, maintained by balancing selection forces and potentially high selfing or inbreeding rates. Moreover, the universality of the non-self recognition genetic systems implies that this type of reproductive isolation might occur in a wide variety of taxonomic groups.
Methods
We used the packages ggplot2 v.3.0.0 (ref. 71), cowplot v.1.0.0 (ref. 72), gridExtra v.2.3 (ref. 73), hexbin v.1.27.3 (ref. 74), rstatix v.0.7.0 (ref. 75) and ggpubr v.0.4.0 (ref. 76) for data visualization and analysis. Animal silhouette figures were taken from http://www.printableparadise.com/printable-animal-silhouettes.html. Most bioinformatics analyses were performed using the workflow manager Snakemake v.5.4.4 (ref. 77).
Fungal material
Including eight previously sequenced strains29, our genomic dataset consisted of 106 P. anserina strains sampled in Wageningen, the Netherlands, and kept at the Laboratory of Genetics of the Wageningen University and Research32,78,79,80. These strains are identified with ‘Wa’ followed by a unique strain ID number (Supplementary Table 1). Most of the strains were obtained between 1991 and 2010 by isolating a single spore from a fruiting body (perithecium) in herbivore dung. Each spore was grown and selfed one or two times to obtain sexual spores that were stored at −80 °C. We further expanded the collection by sampling during 2016 and 2017 (see below) but the strains from 2017 were not sequenced. In addition, we included whole genome sequences of the P. anserina French strains S, Y and Z sampled in 193781,82, the strains CBS433.50 (Canada, 1944), CBS455.64m (Switzerland, 1964), TG (probably from France; see ref. 29), as well as the Podospora pauciseta strain CBS 333.63 (Argentina, 1963). All strains that do not belong to the Wageningen collection have been selfed in the laboratory an unknown, but presumably large, number of times. For strains collected before 2016, we grew a new dikaryotic strain from the frozen stock, selfed it and isolated self-sterile monokaryotic (haploid) spores for sequencing and crosses (Supplementary Fig. 15). From 2016 and 2017, the monokaryons were isolated directly from the first laboratory selfing event. Notice that while the artificial selfing prevents an accurate assessment of heterozygosity, it should not alter the overall diversity in the collection. The mating type of the monokaryon isolates was determined by crossing with tester strains and annotated with a ‘+’ or ‘−’ sign.
Sampling of new P. anserina strains
We collected dung of herbivores (horse, sheep, cow and rabbit) around Wageningen during the months of September and October of 2016 and 2017. Small portions of dung were placed on top of wet filter paper on 9 cm plastic Petri dishes and incubated at room temperature for 2 weeks. Big pieces of dung (mostly from horse) were further fragmented into three to five replicate plates. Humidity was kept by adding a few drops of water (2016) or by keeping the filter paper on top of water agar (2017). After ~10 d of incubation, we observed the emergence of fruiting perithecia resembling P. anserina in 2 and 30 of the dung samples for 2016 and 2017, respectively (replicate plates were counted as one). Individual perithecia were recovered from the dung and placed on a water agar plate covered by an NC 45 membrane filter of Schleicher & Schuell. Using a sterilized needle, we opened the fruiting body, confirmed four-spored asci and proceeded to recover the four spores of a single ascus whenever possible. The single spores were germinated for up to 2 weeks on a plate of PASM2 media with 5 g l−1 ammonium acetate added29. At 2 d postgermination, mycelia was transferred to PASM0.2 plates, incubated for 3–5 d and stored. Only one spore per perithecium was used in all tests (all viable siblings were kept as a backup in the Wageningen collection). In total we sampled 4 (2016) and 68 (2017) strains, from which 5 are probably members of the sibling species P. comata (Supplementary Table 2). No strains were recovered from cow dung. During sampling in 2017, we observed spore killing in the asci of one fruiting body. We defined the identity of this spore killer in strain Wa224 as Psk-7 (containing Spok2, Spok3 and Spok4) on the basis of previously described methodology29,80.
Culture and crossing conditions
Standard methods for growth and manipulation of P. anserina used here are described in the Podospora Genome project homepage (http://podospora.i2bc.paris-saclay.fr/) and in ref. 29. Briefly, strains were grown on Petri dishes with either DO or Henks Perfect barrage (HPM) medium29. Crosses were done either by confronting monokaryotic strains growing together in the same plate or by spermatization. In the latter, mycelia of monokaryotic female parents are fertilized with a suspension of spermatia (microconidia) from a monokaryotic strain used as male parent. Likewise, vegetative compatibility was assessed through barrage formation upon confrontation. Barrage was assessed against a source of light. All cultures were kept at 27 °C with 70% humidity for a 12:12 light:dark cycle. Monokaryotic strains were stored on plates with PASM0.2 media, a calorie-restricted PASM2 recipe83 with 0.2% glucose instead of 2% (PASM0.2) to delay senescence of the isolates.
RI group phenotypic assignment
Wild strains of P. anserina were isolated from horse and sheep dung from Wageningen in 2017 (Supplementary Table 2). Multiple strains could be isolated from the same dung sample. To determine if these isolates represented unique strains or separate isolations of the same clone, all isolates from the same dung sample were confronted against each other under standard conditions on HPM medium. Formation of a barrage between strains was used to indicate that the isolates represented unique genets. If no barrage was formed, isolates were considered to represent the same strain. To confirm that the isolates represented strains of P. anserina, they were confronted against monokaryotic tester strains Wa63 (RV1) and S (rV). New strains can be assigned to RI groups by evaluating the production of perithecia, whereby strains of the same RI group produce more perithecia with each other than to the other RI group. Some strains produce only few perithecia to one or both tester strains and so could not be confidently assigned to an RI group.
Clustering analysis of mating compatibility
Mating compatibility data between the strains of the Wageningen collection produced by ref. 39 was coded into a distance matrix according to the following scheme: mature fruiting bodies produced by both mating partners with or without the presence of ascospore abortion—1; mature fruiting bodies produced by one of the mating partners with or without the presence of ascospore abortion and perithecial abortion observed in the other partner—2; mature fruiting bodies produced by one of the mating partners with or without the presence of ascospore abortion and no perithecia observed on the other partner—3; perithecial abortion observed in both mating partners—4; perithecial abortion observed in one partner of the cross and no perithecia observed in the other partner—5; no perithecia observed on either mating partner—6. The resulting non-symmetric distance matrix (Supplementary Table 3) was transformed into a Gower’s dissimilarity matrix using the R v.3.5.1 function daisy from the package cluster v.2.0.7.1. The optimal number of clusters was determined according to maximum average silhouette width as determined by function pam and the Gap statistic84 also from the cluster package.
DNA extraction and Illumina paired-end sequencing
Growth and DNA extraction followed ref. 29. Briefly, monokaryotic isolates of each strain were grown on PASM0.2 plates covered with cellophane, from which 80–100 mg of mycelium were recovered for extraction. Whole genome DNA was extracted with the ZR Fungal/Bacterial Microprep kit (Zymo; www.zymoresearch.com). Library preparation and sequencing with the HiSeq X Illumina technology (150 base pair (bp) long reads) was done at the SNP and SEQ Technology platform (SciLifeLab). For PCR amplification of het-r (see below), the mycelium was first homogenized in the Mini-Beadbeater (Biospec Products) for 30 s at 25 Hz previous to extraction.
Read mapping and variant calling
We detected adaptors from the Illumina reads using cutadapt v.1.13 (ref. 85) and removed them using Trimmomatic 0.36 (ref. 86) with the following options: ILLUMINACLIP:adaptors.fasta:1:30:9 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:30. We used the filtered pairs with both forward and reverse reads for downstream analysis. As a reference genome we used the second version of the strain S+ assembly45 available in Joint Genome Institute MycoCosm website (https://genome.jgi.doe.gov/programs/fungi/index.jsf) as Podan2. Short-read mapping was done with BWA v.0.7.1787 and PCR duplicates were marked with Picard v.2.19.0 (https://broadinstitute.github.io/picard/). We used HaplotypeCaller of the Genome Analysis Toolkit (GATK) v.4.1.1 (ref. 88) for indel realignment and variant discovery across all (haploid) samples simultaneously, following the GATK Best Practices recommendations89. For subsequent analyses we used only the SNP data (that is, the indels were excluded). Data associated with the mitochondrial scaffold were discarded. VCF file manipulation was done with VCFtools v.0.1.16 (ref. 90) and BCFtools v.1.9 (ref. 91). Visual inspection of the mapped reads revealed low levels (<10% of the total depth of coverage, typically 1–3%) of index-hopping in some samples92. Because the sequenced strains are haploid and have a high read coverage (>80×), the low level of contaminating reads did not affect base calling.
PCA of SNP data
From the SNPs obtained with GATK, we filtered out all sites with the following criteria: QD < 2.0, FS > 60.0, MQ < 40.0, QUAL < 30.0, SOR > 3.0, ReadPosRankSum < −8.0. We also removed all sites that overlapped with repeated elements as detected by RepeatMasker v.4.0.7 (https://www.repeatmasker.org/) with the library of ref. 29. From the remaining SNPs we only retained those sites without any missing data (hereafter referred to as the ‘high-quality’ SNP set). We further reduced the high-quality SNP set by filtering out SNPs with minor allele frequency (MAF) <1% and used the packages gdsfmt v.1.20.0 and SNPrelate v.1.18.0 (ref. 93) in R v.3.5.1 to produce a PCA of the full genome and of each chromosome. A Snakemake pipeline (SNPpop.smk) is available at https://doi.org/10.5281/zenodo.6323682.
This analysis included all samples sequenced, as at least the French strain S also has mating data available to compare it with. However, using only the Wageningen samples leads to similar results. The two samples with intermediate values of PC1 (next to the line at PC1 = 0) in Fig. 2a (Wa60+ and Wa61−), have a V1 allele (that is, they belong to RI group RV1) but are recombinant with the RI group rV for the linked sites surrounding the het-v locus. Removing these two samples had no effect on downstream analyses.
Population genetic analyses
To investigate the genetic diversity of P. anserina, we used the R package PopGenome v.2.6.1 (ref. 94) to estimate the following population genetic parameters: the average pairwise nucleotide diversity within a population (π) and between two populations (Dxy) (ref. 95), Watterson’s theta (θW), the Tajima’s D statistic 96 and the fixation index Fst (refs. 97,98). All analyses were done on windows of 10 kb with steps of 1 kb. Of note, standard VCF files only contain sites that are variable but do not specify sites with missing data that might or might not be invariant. Since ignoring missing data can artificially alter the parameters estimates (for example, some sites that are invariant in the VCF file could actually just be missing data, which would lower estimates of π if the size of the window is not corrected), we used the raw SNP set produced by GATK and we marked explicitly the sites that had either too high or too low coverage as missing data, under the assumption that coverage deviations relate to mis-mapping and repeated elements. To do so, we calculated the depth of coverage distribution of each individual sample and recorded ‘bad’ sites, which were below the 25% quantile or above the 98.5% quantile (thresholds based on preliminary analyses and manual evaluation of reads mapping to the reference genome) using the package vcfR v.1.8.0 (ref. 99) and BEDtools v.2.28.0 (ref. 100). We then intersected regions with ‘bad’ coverage across all individuals with the RepeatMasker annotation above. All sites overlapping with such ranges were set as missing data explicitly in the VCF file and were excluded from the total window length when calculating π and θW. Windows with final length <5 kb were set as missing data. To assess the significance of the Fst values across the genome we randomly rearranged all samples into groups of the same size as the two RI groups (without replacement) and recalculated Fst per window 1,000 times to create a distribution. The location of centromeres was inferred as in ref. 29 on the basis of drops in GC content. A Snakemake pipeline (DiversityStats.smk) is available at https://doi.org/10.5281/zenodo.6323682.
We found that the overall diversity in all sampled strains (Supplementary Table 1) is very low (π = 0.000497). Still, we restricted the analyses to only the Wageningen strains (π = 0.000492) to ensure the patterns correspond to a single population.
LD was calculated using the squared allele frequency correlations (r2) (refs. 101,102). We calculated chromosomal LD decay by combining the methodology of ref. 103 and ref. 104. Specifically, for each chromosome we randomly sampled 30 × 50 kb long windows and extracted all SNPs present in the high-quality SNP with singletons removed. We used VCFtools to calculate r2 for pairs of loci within each window (–hap-r2). We evaluated the decay of LD with distance using a nonlinear regression105 following equation 1 of ref. 106, which assumes a recombination-drift equilibrium model and low levels of mutation. This equation has a single coefficient C equivalent to the product of the distance between SNPs in bp and the effective population recombination rate ρ. This rate, in turn, can be estimated as ρ = 4Nec, where Ne is the effective population size and c is the recombination fraction between sites105,106. We obtained least-squares estimates of ρ by fitting the nls function from the base R104,107. As additional visualization for the general trend in LD decay, we divided the r2 values into distance intervals of 1 kb and computed the mean r2 for each, plotted with a generalized additive mode smoothing (gam method in ggplot2). To find localized patterns of LD along chromosomes, we calculated r2 between pairs of SNPs as above but with a MAF > 0.02 and subsampled so that no two sites are closer than 1 kb of each other (–thin 1000). We then plotted an LD heatmap within each chromosome, or within and between 1 Mb of chromosomes 2 and 5, centred around het-r and het-v, respectively.
We found that the LD decay of chromosome 4 is, on average, much slower than in other chromosomes (Supplementary Fig. 3). This seems to be due to the presence of large haplotypes in both subtelomeric regions and around the het-e locus, which also show very positive Tajima’s D values (Supplementary Fig. 2). When these areas are removed (retaining everything between the positions 1 Mb and 3.5 Mb), the LD decay of chromosome 4 resembles that of other chromosomes (Supplementary Fig. 3).
To assess genome-wide LD with the het-v locus, we calculated r2 between het-v and other markers, either additional genotyped het genes or high-quality biallelic SNPs with a MAF > 0.02 (that is, the variant is present in at least two individuals). The SNPs were processed using the v.1.10.0 of the vcfR package99. As expected given random chromosomal segregation, het-v in chromosome 5 has very low linkage levels (r2 < 0.01) with genotyped het genes in different chromosomes (het-z and het-q). Still, we found a very strong association with het-r in chromosome 2 (r2 = 0.926) as hypothesized due to their interaction but also a moderate association with het-s in chromosome 3 (r2 = 0.246). The latter value, however, falls within the distribution of r2 estimates between het-v and SNPs along the genome in general (Supplementary Fig. 14b).
Cloning and characterization of het-v
Deletion strains were obtained by constructing deletion cassettes in which the Escherichia coli hygromycin B phosphotransferase (hph) or the Streptomyces noursei nourseothricin resistance (nat1) genes were inserted between ~500–700 bp PCR fragments corresponding to the flanking sequences of the region of interest (position of the flanking regions is given Supplementary Fig. 8). The deletion cassettes were then used to transform PaKu70::ble strains derived from the s strain108, which is of the V phenotype. The rV, rV1 and RV1 strains used for phenotypic testing are backcrosses of the respective alleles into an s strain background44. Mutants of the catalytic site residues of HET-Va Y233 were obtained by directed mutagenesis. Homology modelling of the SET and Rubisco domains of HET-Va was done using the HHPred server and Modeller at https://toolkit.tuebingen.mpg.de109. Structure models were visualized with CCP4MG110. Sequence comparison in Hidden Markov model searches were performed with HHMER111 (http://hmmer.org/) and HHPred112.
Positional cloning of het-v
Since het-v was known to be located on the left arm of chromosome 5, we created genetic markers in that region and analysed their linkage to het-v (Fig. 3b and Supplementary Fig. 8). The nat1 marker was introduced in the idi2 gene (Pa_5_4020) located on the left arm of chromosome 5. The nwd3 gene (Pa_5_3370) is also located on the left arm of chromosome 5 and displays two alleles, nwd3-1 and nwd3-2, that can be distinguished by the number of WD40 repeats. A V1 nwd3-2 × V nwd3-1 idi2::nat1 cross was set up and 116 homokaryotic progeny were isolated and phenotyped for nourseothricin resistance, het-v-incompatibility type in barrage tests and genotyped for repeat length polymorphism at nwd3 by PCR. Percentages of second division segregation (see ref. 29 for terminology) were 24%, 49% and 51%, respectively, for idi2, nwd3 and het-r. Out of 103 progeny, 12 showed recombination between idi2::nat1 and V, a single one showed recombination between nwd3 and V and 11 (10.7%) showed recombination between idi2 and nwd3. The physical distance between idi2 and nwd3 is 244 kb, leading to an estimated 1% of recombination for 22.8 kb in that region, suggesting in turn that V is located within ~20 kb (~1% recombination) from nwd3. A 30 kb region spanning the tentative position of V between Pa_5_3300 and Pa_5_3180 was thus amplified and sequenced in a V1 strain. Polymorphism between V and V1 strains was only detected in Pa_5_3200 gene. Deletion of this gene in the V background did not affect V incompatibility and thus Pa_5_3200 is not allelic to V. Because no candidate for V was identified in this region, we set out to obtain a genetic marker that would flank V on the centromere distal side. To that end, the hph marker was introduced between Pa_5_12920 and Pa_5_12915 located at ~165 kb centromere distal to nwd3 and ~115 kb from Pa_5_3180 (the marker was termed hph115). In 124 progeny of a V hph115 × V1 no recombinants between hph115 and V were obtained suggesting tight linkage between the marker and V (but preventing the determination of the relative order of nwd3, V and hph115).
Rather than further increasing the number of analysed progeny (to determine the relative position of nwd3, V and hph115), we reasoned that the position of the het-v gene relative to the hph115 marker could be determined by deleting the 113 kb sequence spanning the region from Pa_5_3180 to the insertion site of the hph115 marker. If V is centromere proximal to hph115, this deletion should affect het-v incompatibility. This region containing 28 annotated genes (Pa_5_3170 to Pa_5_12915) was deleted and replaced by the hph gene (Supplementary Fig. 8). An rV strain was transformed with the deletion cassette and four homokaryotic transformants were obtained. Correct insertion of the deletion cassette was verified by PCR. The transformants deleted for the 113 kb region lost the barrage reaction to rV1. Instead, these transformants now produced a barrage reaction to rV (Fig. 3c). Thus, these transformants lost the V phenotype and at the same time acquired the V1 phenotype, rather than becoming neutral. The het-v locus is thus centromere proximal to hph115 located within the 113 kb region spanning Pa_5_3170 to Pa_5_12915.
Next, we constructed two smaller deletions (77 kb and 35 kb long), further subdividing the 113 kb region containing het-v (Fig. 3b and Supplementary Fig. 8). Deletion of the 77 kb region between Pa_5_12915 and Pa_5_12690 led to transformants displaying the V1 phenotype (it produced a barrage reaction to V). Transformants bearing the 35 kb deletion remained V (it produced a barrage reaction to V1). The same strategy was repeated with deletions Δ12915-12820 and Δ12810-12690 (Supplementary Fig. 8). Transformants carrying the Δ12810-12690 deletion showed the V1 phenotype, those carrying the Δ12915-12820 deletion remained V. Attempts were made to further subdivide the region spanning Pa_5_12810 to Pa_5_12690 but the expected Δ12810-12760 and Δ12750-12690 deletions could not be obtained even after several trials, suggesting that the chromosomal region is recalcitrant to integration of the deletion cassette.
Our results suggest that the deletion of V leads to the V1 phenotype. Accordingly, earlier genetic investigation failed to recover null mutants for V; all mutants recovered for the loss of the V phenotype acquired simultaneously the V1 phenotype42. It also shows that the 113 kb region does not contain any essential genes, consistent with the fact that chromosome 5 appears enriched for contingency genes58.
Identifying the exact ORFs defining het-v
When examining the 113 kb region in the strain Y (of V1 compatibility type), we found that five ORFs present in the reference strain S (V) were replaced by a cluster of transposable elements of the discoglosse (Tc1/mariner-like), atelopus (Copia-Ty1) and crapaud (Gypsy-Ty3) types (see ref. 45 for nomenclature). To determine which of the five ORF(s) within this region confer the V phenotype, we turned to a complementation cloning approach using the Δ12810-12690 V1 strain as a recipient. The region spanning the five ORFs was amplified in a V strain as two PCR fragments, termed a (3.4 kb, corresponding to Pa_5_12750, Pa_5_12740 and Pa_5_12730) and b (5.7 kb, corresponding to Pa_5_12720 and Pa_5_12710) (Fig. 3b and Supplementary Fig. 8). The a and b fragments were used to transform a V1 recipient strain (Δ12810-12690). Ten transformants were obtained with fragment a but none produced a barrage reaction to rV1 nor RV1. From the 18 transformants obtained with fragment b, 14 produced a barrage to rV1 and RV1 and not to rV (they acquired the V phenotype). The remaining four produced no barrage to rV1 or to RV1 (still display the V1 phenotype). Hence, the fragment containing Pa_5_12720 and Pa_5_12710 confers the V phenotype to a V1 strain and thus corresponds to het-v.
Separate roles of Pa_5_12710 (het-Vb) and Pa_5_12720 (het-Va) in het-v incompatibility
The genes het-Va and het-Vb were amplified separately and transformed into the same V1 recipient strain (Supplementary Table 4). Transformants with het-Va alone produced a barrage to RV1 but not to rV1 nor to rV. Transformants with het-Vb alone produced a barrage to rV but not to rV1 or RV1. We conclude from these experiments that het-Va and het-Vb together are required for incompatibility to V1 while het-Va alone determines incompatibility to R.
Analysis of sequences of the proteins encoded by het-Va and het-Vb
Gene het-Va encodes a 469 amino acid long protein with an N-terminal SET lysine methyltransferase domain (pfam 00856), followed by a Rubisco LSMT substrate-binding domain (pfam 09273) and a C-terminal MYND zinc-binding domain (01753) (Fig. 3d and Supplementary Fig. 9). It is homologous to an N. crassa protein that was annotated SET-9 in a systematic description of methyltransferases in that species113. Such SET-domain proteins are predicted lysine methyltransferases of histone and non-histone targets. The best BLASTp and HHPred hits of het-Va in particular are non-histone methyltransferases (last checked on 8 November 2021). Homology modelling to the human SET6D (pdb 3QXY) and Rubisco LSMT protein from Pisum sativum (pdb 2H2J) identified Y233 of Pa_5_12720 as the catalytic tyrosine114. Hence, we generated two point mutants, Y233A and Y233F, which confirmed that the methyltransferase activity of this gene is required for V1/V incompatibility but dispensable for R/V incompatibility (main text). In addition, the two point mutants display a slightly different phenotype: Y233F leads to loss of the barrage reaction to the V tester (and in that sense resembles the wild-type allele), whereas Y233A does not, consistent with the fact that the Y to F substitution is more conservative than Y to A. Homologues of het-Va (displaying the same domain architecture with a SET/Rubisco/MYND domains) were found only in filamentous fungi in a limited number of species (125 blast hits with e-value <10−50 in the JGI fungal database with 2027 genome entries on 5 May 2021).
Gene het-Vb encodes a 771 amino acid long protein with a N-terminal domain (residue 1 to ~140) showing remote similarity with the pore-forming domain of the mammalian MLKL pseudo-kinase involved in execution of necroptotic cell death115. The fungal HeLo and HeLo-like domains involved in the induction of cell death by membrane permeation and in [Het-s]-mediated incompatibility also show homology to MLKL50. The N-terminal region (~535–675) of the het-Vb encoded protein shows homology to TUDOR domains. TUDOR domains are beta-sheet rich globular domains often repeated in tandem and involved in methyl binding49. These low-level homologies were detected using the HHpred server. Gene het-Vb homologues were found only in filamentous fungi in a limited number of species (63 blast hits with e-value <10–50 in the JGI fungal database with 2027 genome entries on 5 May 2021). In a number of species, homologues of both het-Va and het-Vb were found.
Genotyping of the het genes
To genotype their allorecognition genes, all samples with Illumina data were assembled de novo using SPAdes v.3.12.0 (ref. 116) with the k-mers 21, 33, 55, 77 and the --careful option. When available for a given strain, we used the long-read assemblies produced in refs. 29,117 instead. We used BLAST searches to extract the genes defining het-z48, het-s118, het-c31 and het-q119 from all assemblies and we manually assigned each sequence to the corresponding functional allele. Notice that genotyping is not yet possible for the remaining het genes of Podospora (Supplementary Fig. 1). Most variants of het-d and het-e have not been characterized phenotypically, while the gene(s) responsible for het-b have not been cloned yet. We found from sequence alignments of the P. anserina stains with long-read data that there is very little sequence variation within the V and V1 allele classes of the het-v locus.
The Illumina assembly of genes belonging to the HNWD family36,59 proved to be unreliable whenever there were more than four WD40 repeats present (on the basis of comparisons with long-read data assemblies and visual inspections of read mapping). Hence, we followed the PCR protocol of ref. 59 to amplify the WD40 domain of het-r. We used the primers A_F 5′-GCACCGGTTGGCAGTCTGG-3′ and B_R 5′-CCAGGCCCTTCTCGTGTTAGG-3′ in 25 µl of PCR reaction containing 1× Phusion Green GC Buffer (Thermo Scientific), 200 µM of each dNTPs, 0.5 µM of each primer and 0.02 U µl−1 of Phusion High-Fidelity DNA Polymerase (Thermo Scientific). The cycle condition was as follows: an initial denaturation step at 98 °C for 30 s, then 28 cycles each with denaturation at 98 °C for 10 s, annealing at 68 °C for 30 s and elongation at 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. Amplified products were separated in 1.2% agarose gel containing GelRed (Biotium). GeneRuler Express DNA Ladder and GeneRuler 100 bp Plus DNA Ladder (Thermo Scientific) were used as size markers. The number of repeats in the WD40 domain of each strain was estimated on the basis of the size of the WD40 amplicons assuming that one WD40 repeat corresponds to PCR product of about 126 bp.
Since the order and sequence of the WD40 repeats are also important for allele specificity, we selected a subsample of 23 strains for Sanger sequencing (Supplementary Table 1). PCR products were cleaned with Illustra ExoProStar 1-Step enzyme (GF HealthCare), prepared for sequencing with BigDye Terminator v.3.1 Cycle Sequencing and BigDye XTerminator Purification Kits (Applied Biosystems) and sequenced on an ABI 3730 XL machine using two pairs of nested primers (Supplementary Table 5) from ref. 59. We used the Geneious 10.1.3 software (https://www.geneious.com) to align the resulting sequences to the long-read data references to confirm allelic identity. The WD40 repeat structure of the strain Wa63 is considered to be representative of het-R; all other allelic variants are considered to be het-r36. The Sanger sequencing results were consistent with the PCR method.
We reasoned that if the RI groups do not mix in nature, we would encounter a strong deficit of recombinant individuals (rV1). This is a conservative approach to test for reproductive isolation because: (1) errors in the genotyping method above are more likely to favour an r allele since there are many more ways to produce the mutant version (r) rather than finding exactly 11 WD40 repeats in a specific order (R); and (2) spontaneous vegetative mutations are known to occur, probably breaking R into an r allele36,120. Hence, genotyping biases would increase the number of individuals genotyped as rV1.
In the case of the 2017 strains, we genotyped het-v by PCR using a primer pair that specifically amplifies a 4,144 bp region of the V locus (oliVV1: GTGGGAACGATGGAGGGAGAG and oliV: GCGTTCGTCTCAGCAATCTTGAG) and a primer pair that specifically amplifies a 2,177 bp region of the V1 locus. We used the Q5 High-Fidelity DNA Pol kit (NEB) according to the manufacturer’s recommendation. Annealing temperatures and elongation time were 69 °C and 3 min for the V amplification and 66 °C and 2 min for the V1 amplification.
We searched for both het-r and het-v in related species of the P. anserina species complex65 with whole genome sequence data available29,117,121. We used BLAST searches and manual inspection of synteny to assess presence and orthology. We found that the ortholog of het-r is in all species but no strain showed the exact arrangement of 11 repeats found in the P. anserina R allele. While these could correspond to the r allele, further testing and population sampling is needed. In the case of het-v, we only found het-Va and het-Vb in three other strains, representing two species. As the het-v haplotype found in the P. pauciseta strain CBS237.71 was almost identical in sequence to that of P. anserina (suggesting introgression), we chose four genes flanking the het-v haplotype and produced maximum likelihood genealogies of said genes using IQ-TREE v.1.6.8 (refs. 122,123) with extended model selection (-m MFP) and 100 standard bootstrap pseudoreplicates. We kept only six P. anserina strains as representatives of the two RI for clarity within each gene alignment.
To evaluate the presence of the het-v genes in species of Neurospora, the sequences of het-Va and het-Vb were used as a BLAST query against the population datasets from refs. 124,125,126. The custom script query2haplotype v.1 (available at https://github.com/SLAment/Genomics/blob/master/BLAST/query2haplotype.py) was used with parameters -s 100 -e 1 and -f 100. An absence of hits was taken as evidence that that gene is not present in that genome and those with partial hits were considered fragments. The annotation for the gene models of N. crassa and N. tetrasperma (FungiDB, https://fungidb.org/fungidb/) place unsupported introns into the sequences to avoid premature stop codons. We assume that the intron structure matches that of P. anserina as this holds true for the supported introns and for the distantly related sequences from Magnaporthe oryzae. Given this rational, sequences with premature stop codons were assumed to represent putative pseudogenes.
Genotyping of the Spok genes
The presence or absence of Spok2, Spok3 and Spok4 was defined first by BLAST searches as for the het genes above. Whenever Spok3 and Spok4 were present in a sample, either only one gene was assembled (often Spok4) or both were fragmented into multiple small scaffolds. The latter was due to tracks of gene conversion between them29 that complicate the assembly graph. Hence, we mapped the reads of samples with more than one Spok to a reference that has all three Spok types (Wa87+ from ref. 29). Manual inspection of the mapped reads allowed us to distinguish the number and type of Spok genes, as well as to discard false positives due to low levels of index-hopping.
Individual-based simulation of the het-r/het-v interaction in P. anserina
We use SLiM v.3.3.2 (ref. 60) to create individual-based simulations that take into account the genetic architecture of the het-v and het-r genes as well as detailed features of the Podospora life cycle. See Supplementary Methods for details. The SliM and R scripts used to generate and analyse the simulations can be found at https://doi.org/10.5281/zenodo.6323682
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Whole genome sequencing was deposited in NCBI SRA under BioProject PRJNA743020. All other data are available as Supplementary Tables.
Code availability
All custom code is available at Zenodo127 with the identifier https://doi.org/10.5281/zenodo.6323682
References
Nydam, M. L. & DeTomaso, A. W. Creation and maintenance of variation in allorecognition loci: molecular analysis in various model systems. Front. Immunol. 2, 79 (2011).
Rimer, J., Cohen, I. R. & Friedman, N. Do all creatures possess an acquired immune system of some sort? BioEssays 36, 273–281 (2014).
Grice, L. F. & Degnan, B. M. in Evolutionary Transitions to Multicellular Life. Advances in Marine Genomics Vol. 2 (eds Ruiz-Trillo, I. & Nedelcu, A.) 395–424 (Springer, 2015).
Apanius, V., Penn, D., Slev, P. R., Ruff, L. R. & Potts, W. K. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 37, 75–120 (2017).
Glémin, S. et al. Balancing selection in the wild: testing population genetics theory of self-incompatibility in the rare species Brassica insularis. Genetics 171, 279–289 (2005).
Richman, A. Evolution of balanced genetic polymorphism. Mol. Ecol. 9, 1953–1963 (2000).
Tiffin, P. & Moeller, D. A. Molecular evolution of plant immune system genes. Trends Genet. 22, 662–670 (2006).
Wu, J., Saupe, S. J. & Glass, N. L. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl Acad. Sci. USA 95, 12398–12403 (1998).
Eizaguirre, C., Yeates, S. E., Lenz, T. L., Kalbe, M. & Milinski, M. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329 (2009).
Eizaguirre, C., Lenz, T. L., Traulsen, A. & Milinski, M. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol. Lett. 12, 5–12 (2009).
Andreou, D., Eizaguirre, C., Boehm, T. & Milinski, M. Mate choice in sticklebacks reveals that immunogenes can drive ecological speciation. Behav. Ecol. 28, 953–961 (2017).
Bernet, J. Les systèmes d’incompatibilité chez le Podospora anserina. C. R. Hebd. Seances Acad. Sci. D. 265, 1330–1333 (1967).
Pinan-Lucarré, B., Paoletti, M. & Clavé, C. Cell death by incompatibility in the fungus Podospora. Semin. Cancer Biol. 17, 101–111 (2007).
Pedro Gonçalves, A. et al. Conflict, competition, and cooperation regulate social interactions in filamentous fungi. Annu. Rev. Microbiol. 74, 693–712 (2020).
Roca, M. G., Kuo, H. C., Lichius, A., Freitag, M. & Read, N. D. Nuclear dynamics, mitosis, and the cytoskeleton during the early stages of colony initiation in Neurospora crassa. Eukaryot. Cell 9, 1171–1183 (2010).
Roper, M., Simonin, A., Hickey, P. C., Leeder, A. & Glass, N. L. Nuclear dynamics in a fungal chimera. Proc. Natl Acad. Sci. USA 110, 12875–12880 (2013).
Caten, C. E. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72, 221–229 (1972).
Debets, A. J. M. & Griffiths, A. J. F. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol. Res. 102, 1343–1349 (1998).
Debets, F., Yang, X. & Griffiths, A. J. F. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr. Genet. 26, 113–119 (1994).
Hartl, D. L., Dempster, E. R. & Brown, S. W. Adaptive significance of vegetative incompatibility in Neurospora crassa. Genetics 81, 553–569 (1975).
Zhang, D. X., Spiering, M. J., Dawe, A. L. & Nuss, D. L. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 197, 701–714 (2014).
Glass, N. L. & Kaneko, I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell https://doi.org/10.1128/EC.2.1.1-8.2003 (2003).
Paoletti, M. & Saupe, S. J. Fungal incompatibility: evolutionary origin in pathogen defense? BioEssays 31, 1201–1210 (2009).
Uehling, J., Deveau, A. & Paoletti, M. Do fungi have an innate immune response? An NLR-based comparison to plant and animal immune systems. PLoS Pathog. 13, e1006578 (2017).
Garnjobst, L. & Wilson, J. F. Heterocaryosis and protoplasmic incompatibility in Neurospora crassa. Proc. Natl Acad. Sci. USA 42, 613–618 (1956).
Glass, N. L. & Kuldau, G. A. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu. Rev. Phytopathol. 30, 201–224 (1992).
Esser, K. in Growth, Differentiation and Sexuality. The Mycota Vol. I (ed. Wendland, J.) 103–131 (Springer International, 2016).
Nieuwenhuis, B. P. S. & James, T. Y. The frequency of sex in fungi. Philos. Trans. R. Soc. B 371, 20150540 (2016).
Vogan, A. A. et al. Combinations of Spok genes create multiple meiotic drivers in Podospora. eLife 8, e46454 (2019).
Grognet, P., Lalucque, H., Malagnac, F. & Silar, P. Genes that bias Mendelian segregation. PLoS Genet. https://doi.org/10.1371/journal.pgen.1004387 (2014).
Bastiaans, E. et al. Natural variation of heterokaryon incompatibility gene het-c in Podospora anserina reveals diversifying selection. Mol. Biol. Evol. 31, 962–974 (2014).
Debets, A. J. M. et al. High natural prevalence of a fungal prion. Proc. Natl Acad. Sci. USA 109, 10432–10437 (2012).
Mir-Rashed, N., Jacobson, D. J., Dehghany, M. R., Micali, O. C. & Smith, M. L. Molecular and functional analyses of incompatibility genes at het-6 in a population of Neurospora crassa. Fungal Genet. Biol. 30, 197–205 (2000).
Muirhead, C. A., Louise Glass, N. & Slatkin, M. Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics 161, 633–641 (2002).
Zhao, J. et al. Identification of allorecognition loci in Neurospora crassa by genomics and evolutionary approaches. Mol. Biol. Evol. 32, 2417–2432 (2015).
Chevanne, D., Saupe, S. J., Clavé, C. & Paoletti, M. WD-repeat instability and diversification of the Podospora anserina hnwd non-self recognition gene family. BMC Evol. Biol. 10, 134 (2010).
Paoletti, M., Saupe, S. J. & Clavé, C. Genesis of a fungal non-self recognition repertoire. PLoS ONE 2, e283 (2007).
Saupe, S., Descamps, C., Turcq, B. & Begueret, J. Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc. Natl Acad. Sci. USA 91, 5927–5931 (1994).
van der Gaag, M. Genomic conflicts in Podospora anserina. PhD thesis, Wageningen Universiteit (2005).
Marcou, D., Picard-Bennoun, M. & Simonet, J.-M. in Genetic Maps (ed. O’Brien, S. J.) 3.92–3.101 (Cold Spring Harbor Laboratory Press, 1982).
Labarère, J., Bègueret, J. & Bernet, J. Incompatibility in Podospora anserina: comparative properties of the antagonistic cytoplasmic factors of a nonallelic system. J. Bacteriol. 120, 854–860 (1974).
Labarère, J. L’Incompatibilité Protoplasmique Chez le Champignon Podospora anserina: Etude Génétique et Biochimique, Relation avec Certains Aspects du Développement et de la Morphogénèse. PhD thesis, Université Bordeaux 2 (1978).
Bernet, J. Mode d’action des gènes de barrage et relation entre l’incompatibilité cellulaire et l’incompatibilité sexuelle chez le Podospora anserina. Ann. Sci. Nat. Bot. 6, 611–768 (1965).
Labarère, J. Propriétes d’un système d’incompatibilité chez le champignon Podospora anserina et intérêt de ce système pour l’étude de l’incompatibilté. C. R. Acad. Sci. 276, 1301–1304 (1973).
Espagne, E. et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9, R77 (2008).
Deleu, C. Analyse Moléculaire du Locus het-s et Etude de l’Expression des Allèles het-s et het-S Responsables d’Incompatibilité Végétative Chez le Champignon Filamenteux Podospora anserina. PhD thesis, Université Bordeaux 2 (1993).
Daskalov, A., Dyrka, W. & Saupe, S. J. Theme and variations: evolutionary diversification of the HET-s functional amyloid motif. Sci. Rep. 5, 12494 (2015).
Heller, J., Clavé, C., Gladieux, P., Saupe, S. J. & Glass, N. L. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl Acad. Sci. USA 115, E2292–E2301 (2018).
Lu, R. & Wang, G. G. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 38, 546–555 (2013).
Daskalov, A. et al. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc. Natl Acad. Sci. USA 113, 2720–2725 (2016).
Jones, J. D. G., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science 354, 6316 (2016).
Meunier, E. & Broz, P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 38, 744–757 (2017).
Orr, H. A. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144, 1331–1335 (1996).
Bomblies, K. & Weigel, D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8, 382–393 (2007).
Bomblies, K. et al. Autoimmune response as a mechanism for a Dobzhansky–Muller-type incompatibility syndrome in plants. PLoS Biol. 5, 1962–1972 (2007).
Krüger, J. et al. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296, 744–747 (2002).
Deng, J. et al. A CC-NBS-LRR gene induces hybrid lethality in cotton. J. Exp. Bot. 70, 5145–5156 (2019).
Bidard, F., Clavé, C. & Saupe, S. J. The transcriptional response to nonself in the fungus Podospora anserina. G3 3, 1015–1030 (2013).
Chevanne, D. et al. Identification of the het-r vegetative incompatibility gene of Podospora anserina as a member of the fast evolving HNWD gene family. Curr. Genet. 55, 93–102 (2009).
Haller, B. C. & Messer, P. W. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36, 632–637 (2019).
Ispolatov, I. & Doebeli, M. Speciation due to hybrid necrosis in plant–pathogen models. Evolution 63, 3076–3084 (2009).
Glémin, S. Surprising fitness consequences of GC-biased gene conversion. I. Mutation load and inbreeding depression. Genetics 185, 939–959 (2010).
Martinossi‐Allibert, I. et al. Invasion and maintenance of meiotic drivers in populations of ascomycete fungi. Evolution 75, 1150–1169 (2021).
Meiklejohn, C. D. et al. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife 7, e35468 (2018).
Boucher, C., Nguyen, T.-S. & Silar, P. Species delimitation in the Podospora anserina/ P. pauciseta/ P. comata species complex (Sordariales). Cryptogam. Mycol. 38, 485–506 (2017).
Kruys, Å., Huhndorf, S. M. & Miller, A. N. Coprophilous contributions to the phylogeny of Lasiosphaeriaceae and allied taxa within Sordariales (Ascomycota, Fungi). Fungal Divers. 70, 101–113 (2015).
Menkis, A., Bastiaans, E., Jacobson, D. J. & Johannesson, H. Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J. Evol. Biol. 22, 1923–1936 (2009).
Coyne, J. A. & Orr, H. A. Speciation (Sinauer Associates, 2004).
Nosil, P., Feder, J. L. & Gompert, Z. How many genetic changes create new species? Science 371, 777–779 (2021).
Nosil, P. & Schluter, D. The genes underlying the process of speciation. Trends Ecol. Evol. 26, 160–167 (2011).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Wilke, C. O. cowplot: Streamlined plot theme and plot annotations for “ggplot2”. R version 1.0.0 (2019).
Auguie, B. gridExtra: Miscellaneous functions for “Grid” graphics. R version 2.3 (2017).
Carr, D. hexbin: Hexagonal binning routines. R version 1.27.3 (2019).
Kassambara, A. rstatix: Pipe-friendly framework for basic statistical tests. R version 0.7.0 (2021).
Kassambara, A. ggpubr: “ggplot2” based publication ready plots. R version 0.4.0 (2020).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Hermanns, J., Debets, F., Hoekstra, R. & Osiewacz, H. D. A novel family of linear plasmids with homology to plasmid pAL2-1 of Podospora anserina. Mol. Gen. Genet. 246, 638–647 (1995).
van der Gaag, M., Debets, A. J. M., Osiewacz, H. D. & Hoekstra, R. F. The dynamics of pAL2-1 homologous linear plasmids in Podospora anserina. Mol. Gen. Genet. 258, 521–529 (1998).
van der Gaag, M. et al. Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserina. Genetics 156, 593–605 (2000).
Belcour, L., Rossignol, M., Koll, F., Sellem, C. H. & Oldani, C. Plasticity of the mitochondrial genome in Podospora. Polymorphism for 15 optional sequences: group-I, group-II introns, intronic ORFs and an intergenic region. Curr. Genet. 31, 308–317 (1997).
Rizet, G. Les phénomènes de barrage chez Podospora anserina. I. Analyse genetique des barrages entre souches S et s. Rev. Cytol. Biol. Veg. 13, 51–92 (1952).
van Diepeningen, A. D., Debets, A. J. M., Slakhorst, S. M. & Hoekstra, R. F. Mitochondrial pAL2-1 plasmid homologs are senescence factors in Podospora anserina independent of intrinsic senescence. Biotechnol. J. 3, 791–802 (2008).
Tibshirani, R., Walther, G. & Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. B 63, 411–423 (2001).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Preprint at bioRxiv https://doi.org/10.1101/201178 (2017).
Van der Auwera, G. A. et al. From FastQ data to high‐confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11.10.1–11.10.33 (2013).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
van der Valk, T., Vezzi, F., Ormestad, M., Dalén, L. & Guschanski, K. Index hopping on the Illumina HiseqX platform and its consequences for ancient DNA studies. Mol. Ecol. Resour. 20, 1171–1181 (2020).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Pfeifer, B., Wittelsbürger, U., Ramos-Onsins, S. E. & Lercher, M. J. PopGenome: an efficient Swiss Army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936 (2014).
Nei, M. & Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269–5273 (1979).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Wright, S. Isolation by distance. Genetics 28, 114–138 (1943).
Wright, S. Evolution in Mendelian populations. Genetics 16, 97–159 (1931).
Knaus, B. J. & Grünwald, N. J. vcfR: a package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53 (2017).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Hill, W. G. & Robertson, A. R. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231 (1968).
Lewontin, R. C. On measures of gametic disequilibrium. Genetics 120, 849–852 (1988).
Hench, K., Vargas, M., Höppner, M. P., McMillan, W. O. & Puebla, O. Inter-chromosomal coupling between vision and pigmentation genes during genomic divergence. Nat. Ecol. Evol. 3, 657–667 (2019).
Marroni, F. et al. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet. Genomes 7, 1011–1023 (2011).
Hill, W. G. & Weir, B. S. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Pop. Biol. 33, 54–78 (1988).
Remington, D. L. et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl Acad. Sci. USA 98, 11479–11484 (2001).
Tusso, S. et al. Ancestral admixture is the main determinant of global biodiversity in fission yeast. Mol. Biol. Evol. 36, 1975–1989 (2019).
El-Khoury, R. et al. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53, 249–258 (2008).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K. & Noble, M. The CCP4 molecular-graphics project. Acta Crystallogr. D 58, 1955–1957 (2002).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37 (2011).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, 244–248 (2005).
Adhvaryu, K. K., Morris, S. A., Strahl, B. D. & Selker, E. U. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell 4, 1455–1464 (2005).
Chang, Y. et al. Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling. Nucleic Acids Res. 39, 6380–6389 (2011).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Vogan, A. A. et al. The Enterprise, a massive transposon carrying Spok meiotic drive genes. Genome Res. 31, 789–798 (2021).
Deleu, C., Clavé, C. & Bégueret, J. A single amino acid difference is sufficient to elicit vegetative incompatibility in the fungus Podospora anserina. Genetics 135, 45–52 (1993).
Clavé, C. et al. Fungal gasdermin-like proteins are controlled by proteolytic cleavage. Proc. Natl Acad. Sci. USA 119, e2109418119 (2022).
Espagne, E., Balhadère, P., Penin, M. L., Barreau, C. & Turcq, B. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics 161, 71–81 (2002).
Hartmann, F. E. et al. Size variation of the nonrecombining region on the mating-type chromosomes in the fungal Podospora anserina species complex. Mol. Biol. Evol. 38, 2475–2492 (2021).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective sochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Corcoran, P. et al. Introgression maintains the genetic integrity of the mating-type determining chromosome of the fungus Neurospora tetrasperma. Genome Res. 26, 486–498 (2016).
Svedberg, J. et al. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat. Commun. 9, 4242 (2018).
Svedberg, J. et al. An introgressed gene causes meiotic drive in Neurospora sitophila. Proc. Natl Acad. Sci. USA 118, e2026605118 (2021).
Ament-Velásquez, S. L. Code of ‘Allorecognition genes drive reproductive isolation in Podospora anserina.’ v.1.0.0. Zenodo https://doi.org/10.5281/zenodo.6323682 (2022).
Acknowledgements
We would like to thank M. Grudzinska-Sterno for valuable assistance with laboratory work, as well as R. Debuchy and M. Sicault-Sabourin for assistance in the positional cloning. Next Generation sequencing was possible thanks to the support of the National Genomics Infrastructure/Uppsala Genome Center on massive parallel DNA sequencing. The computations were performed on resources provided by Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under the projects SNIC 2017/1-567 and SNIC 2019/8-371. We would also like to thank S. Tusso for useful advice and code peer review. Funding was provided by the European Research Council (ERC) with the grant no. ERC-2014-CoG, project 648143 SpoKiGen (H.J.); the Swedish Research Council with grant no. 2015-04649 (H.J.); the Centre National de la Eecherche Scientifique with recurrent funding to UMR 5095 (S.J.S.); and the Anna Maria Lundin’s Travel Grant 2016 and 2017 (S.L.A.-V.).
Funding
Open access funding provided by Uppsala University.
Author information
Authors and Affiliations
Contributions
S.L.A.-V., A.A.V., H.J., S.J.S. and C.C. were responsible for conceptualization. S.L.A.-V., A.A.V., E.B. and S.D.G. undertook data curation. S.L.A.-V., A.A.V., I.M.-A. and S.J.S. completed the formal analysis. H.J., S.J.S. and S.L.A.-V. acquired funding. S.L.A.-V., A.A.V., A.G.-F., E.B., I.M.-A., S.J.S., S.D.G. and C.C. conducted investigations. S.L.A.-V., A.A.V., A.G.-F., I.M.-A. and C.C. were involved in methodology. S.L.A.-V., A.A.V., S.J.S. and H.J. undertook project administration. A.J.M.D. obtained resources. S.L.A.-V. was responsible for software. H.J., S.J.S., A.J.M.D. and M.L. undertook supervision. S.L.A.-V., I.M.-A. and S.J.S. conducted visualization. S.L.A.-V. wrote the orginal draft manuscript. S.L.A.-V., A.A.V., I.M.-A., M.L., H.J., S.J.S. and A.J.M.D. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Jun-Yi Leu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–19, Tables 4 and 6 and captions of Tables 1, 2, 3, 5 and 7.
Supplementary Tables
Supplementary Tables 1, 2, 3, 5 and 7.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ament-Velásquez, S.L., Vogan, A.A., Granger-Farbos, A. et al. Allorecognition genes drive reproductive isolation in Podospora anserina. Nat Ecol Evol 6, 910–923 (2022). https://doi.org/10.1038/s41559-022-01734-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-022-01734-x